|
Oligosaccharyltransferase
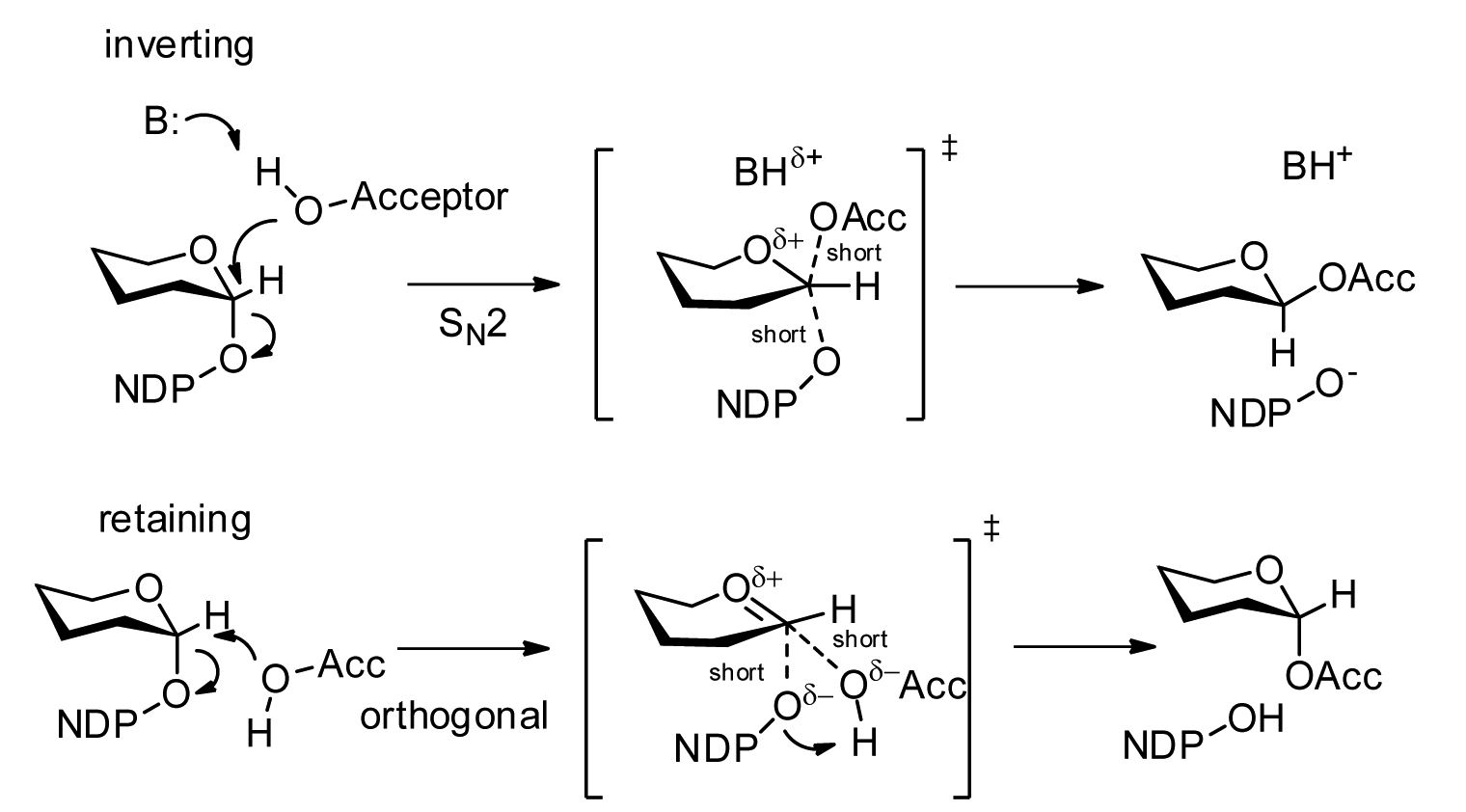
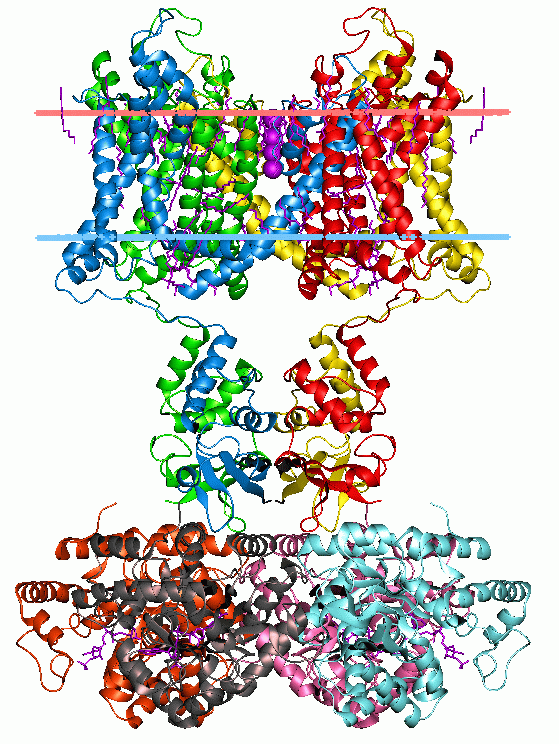
Oligosaccharyltransferase or OST () is a membrane protein protein complex, complex that transfers a 14-sugar oligosaccharide from dolichol to nascent protein. It is a type of glycosyltransferase. The sugar Glc3Man9GlcNAc2 (where Glc=Glucose, Man=Mannose, and GlcNAc=N-Acetylglucosamine, ''N''-acetylglucosamine) is attached to an asparagine (Asn) residue in the sequence Asn-X-Serine, Ser or Asn-X-Threonine, Thr where X is any amino acid except proline. This sequence is called a glycosylation ''sequon.'' The reaction catalyzed by OST is the central step in the Glycosylation#N-linked glycosylation, ''N''-linked glycosylation Metabolic pathway, pathway. Location OST is a component of the translocon in the endoplasmic reticulum (ER) Membrane (biology), membrane. A lipid-linked core-oligosaccharide is assembled at the membrane of the endoplasmic reticulum and transferred to selected asparagine residues of nascent polypeptide chains by the oligosaccharyl transferase Protein complex, co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycosyltransferase
Glycosyltransferases (GTFs, Gtfs) are enzymes ( EC 2.4) that establish natural glycosidic linkages. They catalyze the transfer of saccharide moieties from an activated nucleotide sugar (also known as the "glycosyl donor") to a nucleophilic glycosyl acceptor molecule, the nucleophile of which can be oxygen- carbon-, nitrogen-, or sulfur-based. The result of glycosyl transfer can be a carbohydrate, glycoside, oligosaccharide, or a polysaccharide. Some glycosyltransferases catalyse transfer to inorganic phosphate or water. Glycosyl transfer can also occur to protein residues, usually to tyrosine, serine, or threonine to give O-linked glycoproteins, or to asparagine to give N-linked glycoproteins. Mannosyl groups may be transferred to tryptophan to generate C-mannosyl tryptophan, which is relatively abundant in eukaryotes. Transferases may also use lipids as an acceptor, forming glycolipids, and even use lipid-linked sugar phosphate donors, such as dolichol phosphates in eukaryotic o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Membrane Protein
Membrane proteins are common proteins that are part of, or interact with, biological membranes. Membrane proteins fall into several broad categories depending on their location. Integral membrane proteins are a permanent part of a cell membrane and can either penetrate the membrane (transmembrane) or associate with one or the other side of a membrane ( integral monotopic). Peripheral membrane proteins are transiently associated with the cell membrane. Membrane proteins are common, and medically important—about a third of all human proteins are membrane proteins, and these are targets for more than half of all drugs. Nonetheless, compared to other classes of proteins, determining membrane protein structures remains a challenge in large part due to the difficulty in establishing experimental conditions that can preserve the correct conformation of the protein in isolation from its native environment. Function Membrane proteins perform a variety of functions vital to the sur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amide
In organic chemistry, an amide, also known as an organic amide or a carboxamide, is a compound with the general formula , where R, R', and R″ represent organic groups or hydrogen atoms. The amide group is called a peptide bond when it is part of the main chain of a protein, and an isopeptide bond when it occurs in a side chain, such as in the amino acids asparagine and glutamine. It can be viewed as a derivative of a carboxylic acid () with the hydroxyl group () replaced by an amine group (); or, equivalently, an acyl (alkanoyl) group () joined to an amine group. Common examples of amides are acetamide (), benzamide (), and dimethylformamide (). Amides are qualified as primary, secondary, and tertiary according to whether the amine subgroup has the form , , or , where R and R' are groups other than hydrogen. The core of amides is called the amide group (specifically, carboxamide group). Amides are pervasive in nature and technology. Proteins and important plastics l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Asparagine
Asparagine (symbol Asn or N) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −COO− form under biological conditions), and a side chain carboxamide, classifying it as a polar (at physiological pH), aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it. It is encoded by the codons AAU and AAC. History Asparagine was first isolated in 1806 in a crystalline form by French chemists Louis Nicolas Vauquelin and Pierre Jean Robiquet (then a young assistant). It was isolated from asparagus juice, in which it is abundant, hence the chosen name. It was the first amino acid to be isolated. Three years later, in 1809, Pierre Jean Robiquet identified a substance from liquorice root with properties which he qualified as very similar to those of asparagine, and which Plisson identi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular mass.'' The name is composed of Greek elements '' oligo-'', "a few" and '' -mer'', "parts". An adjective form is ''oligomeric''. The oligomer concept is contrasted to that of a polymer, which is usually understood to have a large number of units, possibly thousands or millions. However, there is no sharp distinction between these two concepts. One proposed criterion is whether the molecule's properties vary significantly with the removal of one or a few of the units. An oligomer with a specific number of units is referred to by the Greek prefix denoting that number, wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
OST4
In molecular biology, OST4 (Dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit 4) is a subunit of the oligosaccharyltransferase complex. OST4 is a very short, approximately 30 amino acids, protein found from fungi to vertebrates. It appears to be an integral membrane protein that mediates the en bloc transfer of a pre-assembled high-mannose oligosaccharide onto asparagine residues of nascent polypeptides as they enter the lumen of the rough endoplasmic reticulum The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum ( .... References {{InterPro content, IPR018943 Protein families Chi, J.H., Roos, J. and N. Dean. (1996) The OST4 gene of Saccharomyces cerevisiae encodes an unusually small protein required for normal levels of oligosaccharyltransferase activity J. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Folding
Protein folding is the physical process by which a protein chain is translated to its native three-dimensional structure, typically a "folded" conformation by which the protein becomes biologically functional. Via an expeditious and reproducible process, a polypeptide folds into its characteristic three-dimensional structure from a random coil. Each protein exists first as an unfolded polypeptide or random coil after being translated from a sequence of mRNA to a linear chain of amino acids. At this stage the polypeptide lacks any stable (long-lasting) three-dimensional structure (the left hand side of the first figure). As the polypeptide chain is being synthesized by a ribosome, the linear chain begins to fold into its three-dimensional structure. Folding of many proteins begins even during translation of the polypeptide chain. Amino acids interact with each other to produce a well-defined three-dimensional structure, the folded protein (the right hand side of the figure), ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Posttranslational Modification
Post-translational modification (PTM) is the covalent and generally enzymatic modification of proteins following protein biosynthesis. This process occurs in the endoplasmic reticulum and the golgi apparatus. Proteins are synthesized by ribosomes translating mRNA into polypeptide chains, which may then undergo PTM to form the mature protein product. PTMs are important components in cell signal transduction, signaling, as for example when prohormones are converted to hormones. Post-translational modifications can occur on the amino acid side chains or at the protein's C-terminus, C- or N-terminus, N- termini. They can extend the chemical repertoire of the 20 standard amino acids by modifying an existing functional group or introducing a new one such as phosphorylation, phosphate. Phosphorylation is a highly effective mechanism for regulating the activity of enzymes and is the most common post-translational modification. Many eukaryotic and prokaryotic proteins also have carbohydra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translation (biology)
In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription (biology), transcription of DNA to RNA in the cell's nucleus (cell), nucleus. The entire process is called gene expression. In translation, mRNA, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later protein folding, folds into an Activation energy, active protein and performs its functions in the Cell (biology), cell. The ribosome facilitates decoding by inducing the binding of Base pair, complementary tRNA anticodon sequences to mRNA codons. The tRNAs carry specific amino acids that are chained together into a polypeptide as the mRNA passes through and is "read" by the ribosome. Translation proceeds in three phases: # Initiation: The ribosome assembles around the target mRNA. The first tRNA is attached a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gunnar Von Heijne
Professor Nils ''Gunnar'' Hansson von Heijne, born 10 June 1951 in Gothenburg, is a Swedish scientist working on signal peptides, membrane proteins and bioinformatics at the Stockholm Center for Biomembrane Research at Stockholm University. Education Gunnar von Heijne graduated 1975 with a Master of Science degree in chemistry and chemical engineering from the Royal Institute of Technology (KTH).Gunnar von Heijne's CV He then became a doctoral student in at KTH, in a research group focussing on and the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multienzyme complexes, in which multiple catalytic domains are found in a single polypeptide chain. Protein complexes are a form of quaternary structure. Proteins in a protein complex are linked by non-covalent protein–protein interactions. These complexes are a cornerstone of many (if not most) biological processes. The cell is seen to be composed of modular supramolecular complexes, each of which performs an independent, discrete biological function. Through proximity, the speed and selectivity of binding interactions between enzymatic complex and substrates can be vastly improved, leading to higher cellular efficiency. Many of the techniques used to enter cells and isolate proteins are inherently disruptive to such large complexes, complicating the task of determining the components of a complex. Examples of protein complexes include the p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |