|
Fas Receptor
The Fas receptor, also known as Fas, FasR, apoptosis antigen 1 (APO-1 or APT), cluster of differentiation 95 (CD95) or tumor necrosis factor receptor superfamily member 6 (TNFRSF6), is a protein that in humans is encoded by the ''FAS'' gene. Fas was first identified using a monoclonal antibody generated by immunizing mice with the FS-7 cell line. Thus, the name Fas is derived from ''F''S-7-''a''ssociated ''s''urface antigen. The Fas receptor is a death receptor on the surface of cells that leads to programmed cell death (apoptosis) if it binds its ligand, Fas ligand (FasL). It is one of two apoptosis pathways, the other being the mitochondrial pathway. Gene FAS receptor gene is located on the long arm of chromosome 10 (10q24.1) in humans and on chromosome 19 in mice. The gene lies on the plus ( Watson strand) and is 25,255 bases in length organized into nine protein encoding exons. Similar sequences related by evolution (orthologs) are found in most mammals. Protein Prev ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cluster Of Differentiation
The cluster of differentiation (also known as cluster of designation or classification determinant and often abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules providing targets for immunophenotyping of cells. In terms of physiology, CD molecules can act in numerous ways, often acting as receptors or ligands important to the cell. A signal cascade is usually initiated, altering the behavior of the cell (see cell signaling). Some CD proteins do not play a role in cell signaling, but have other functions, such as cell adhesion. CD for humans is numbered up to 371 (). Nomenclature The CD nomenclature was proposed and established in the 1st International Workshop and Conference on Human Leukocyte Differentiation Antigens (HLDA), which was held in Paris in 1982. This system was intended for the classification of the many monoclonal antibodies (mAbs) generated by different laboratories around the world against epitopes on the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TIA1
TIA1 or Tia1 cytotoxic granule-associated rna binding protein is a 3'UTR mRNA binding protein that can bind the 5'TOP sequence of 5'TOP mRNAs. It is associated with programmed cell death (apoptosis) and regulates alternative splicing of the gene encoding the Fas receptor, an apoptosis-promoting protein. Under stress conditions, TIA1 localizes to cellular RNA-protein conglomerations called stress granules. It is encoded by the TIA1 gene. Mutations in the ''TIA1'' gene have been associated with amyotrophic lateral sclerosis, frontotemporal dementia, and Welander distal myopathy. It also plays a crucial role in the development of toxic oligomeric tau in Alzheimer's disease. Function This protein is a member of a RNA-binding protein family that regulates transcription and RNA translation. It was first identified in cytotoxic lymphocyte (CTL) target cells. TIA1 acts in the nucleus to regulate splicing and transcription. TIA1 helps to recruit the splicesome to regulate RNA spli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chimeric Antigen Receptor T Cell
In biology, chimeric antigen receptors (CARs)—also known as chimeric immunoreceptors, chimeric T cell receptors or artificial T cell receptors—are receptor proteins that have been engineered to give T cells the new ability to target a specific antigen. The receptors are chimeric in that they combine both antigen-binding and T cell activating functions into a single receptor. CAR T cell therapy uses T cells engineered with CARs to treat cancer. The premise of CAR T immunotherapy is to modify T cells to recognize cancer cells in order to more effectively target and destroy them. Scientists harvest T cells from people, genetically alter them, then infuse the resulting CAR T cells into patients to attack their tumors. CAR T cells can be derived either from T cells in a patient's own blood (autologously) or from the T cells of another, healthy, donor ( allogeneically). Once isolated from a person, these T cells are genetically engineered to express a specific CAR, which programs t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Icahn School Of Medicine At Mount Sinai
The Icahn School of Medicine at Mount Sinai (ISMMS or Mount Sinai), formerly the Mount Sinai School of Medicine, is a private medical school in New York City. It is the academic teaching arm of the Mount Sinai Health System, which manages eight hospital campuses in the New York metropolitan area, including Mount Sinai Hospital and the New York Eye and Ear Infirmary. Mount Sinai is ranked #11 among American medical schools by the 2023 '' U.S. News & World Report''.https://www.usnews.com/best-graduate-schools/top-medical-schools/icahn-school-of-medicine-at-mount-sinai-04072 In 2021, it was ranked 15th in the country for biomedical research and leads the country in research funding from the National Institutes of Health for neuroscience (#2) and genetics (#2). It attracted over $400 million in total NIH funding in 2021. Mount Sinai's faculty includes 23 elected members of the National Academies of Sciences, Engineering, and Medicine and 40 members of the American Society for Cl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tumor Suppressor
A tumor suppressor gene (TSG), or anti-oncogene, is a gene that regulates a cell during cell division and replication. If the cell grows uncontrollably, it will result in cancer. When a tumor suppressor gene is mutated, it results in a loss or reduction in its function. In combination with other genetic mutations, this could allow the cell to grow abnormally. The loss of function for these genes may be even more significant in the development of human cancers, compared to the activation of oncogenes. TSGs can be grouped into the following categories: caretaker genes, gatekeeper genes, and more recently landscaper genes. Caretaker genes ensure stability of the genome via DNA repair and subsequently when mutated allow mutations to accumulate. Meanwhile, gatekeeper genes directly regulate cell growth by either inhibiting cell cycle progression or inducing apoptosis. Lastly landscaper genes regulate growth by contributing to the surrounding environment, when mutated can cause an envir ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
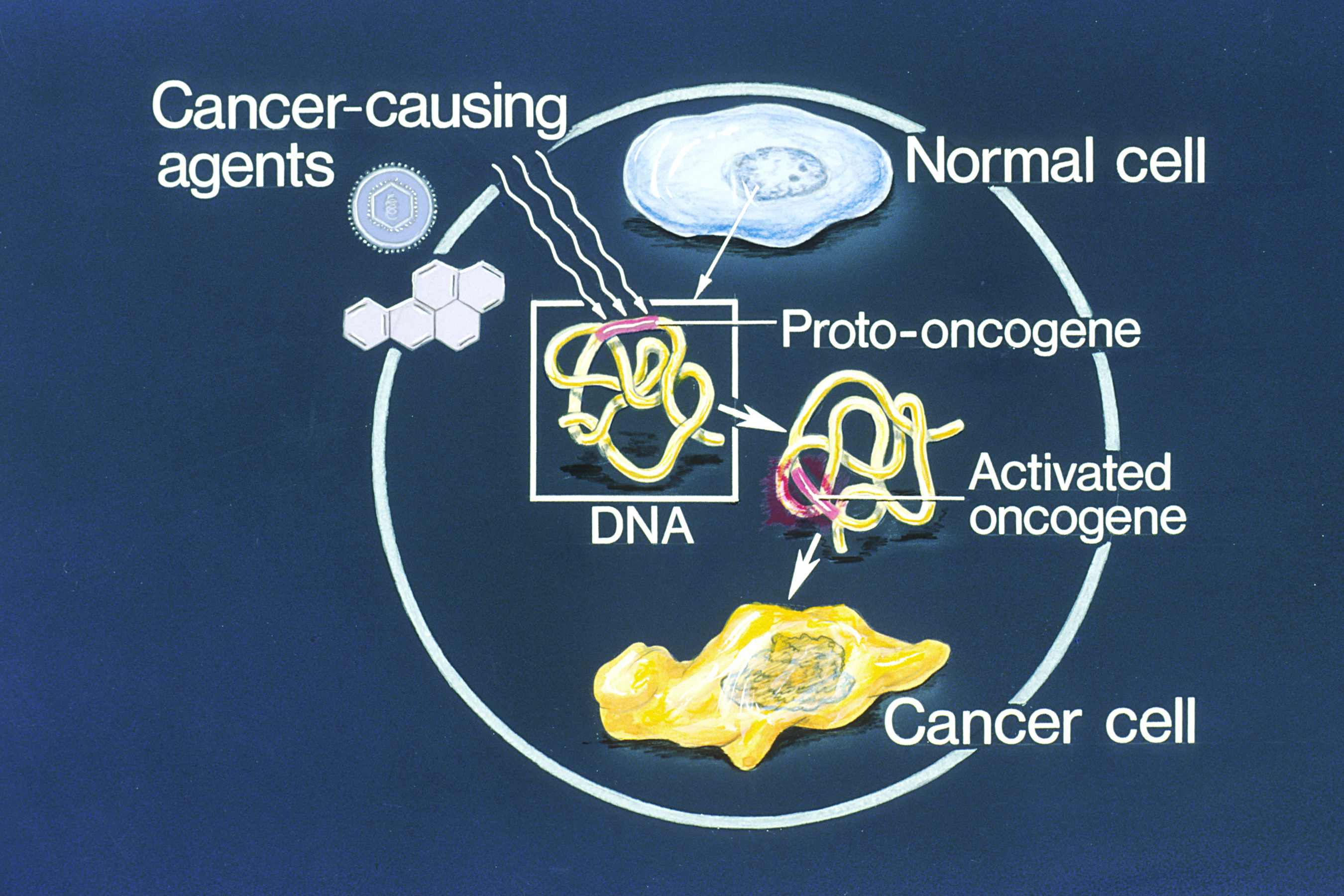
Oncogene
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.Kimball's Biology Pages. "Oncogenes" Free full text Most normal cells will undergo a programmed form of rapid cell death () when critical functions are altered and malfunctioning. Activated oncogenes can cause those cells designated for apoptosis to survive and proliferate instead. Most oncogenes began as proto-oncogenes: normal genes involved in cell growth and proliferation or inhibition of apoptosis. If, through mutation, normal genes promoting cellular growth are up-regulated (gain-of-function mutation), they will predisp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteolytic Cleavage
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion. Proteolysis in organisms serves many purposes; for example, digestive enzymes break down proteins in food to provide amino acids for the organism, while proteolytic processing of a polypeptide chain after its synthesis may be necessary for the production of an active protein. It is also important in the regulation of some physiological and cellular processes including apoptosis, as well as preventing the accumulation of unwanted or misfolded proteins in cells. Consequently, abnormality in the regulation of proteolysis can cause disease. Proteolysis can also be used as an analytical tool for studying proteins in the laboratory, and it may also be used in industry, for example in food proces ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Caspase-8
Caspase-8 is a caspase protein, encoded by the ''CASP8'' gene. It most likely acts upon caspase-3. ''CASP8'' orthologs have been identified in numerous mammals for which complete genome data are available. These unique orthologs are also present in birds. Function The ''CASP8'' gene encodes a member of the cysteine-aspartic acid protease (caspase) family. Sequential activation of caspases plays a central role in the execution-phase of cell apoptosis. Caspases exist as inactive proenzymes composed of a prodomain, a large protease subunit, and a small protease subunit. Activation of caspases requires proteolytic processing at conserved internal aspartic residues to generate a heterodimeric enzyme consisting of the large and small subunits. This protein is involved in the programmed cell death induced by Fas and various apoptotic stimuli. The N-terminal FADD-like death effector domain of this protein suggests that it may interact with Fas-interacting protein FADD. This protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
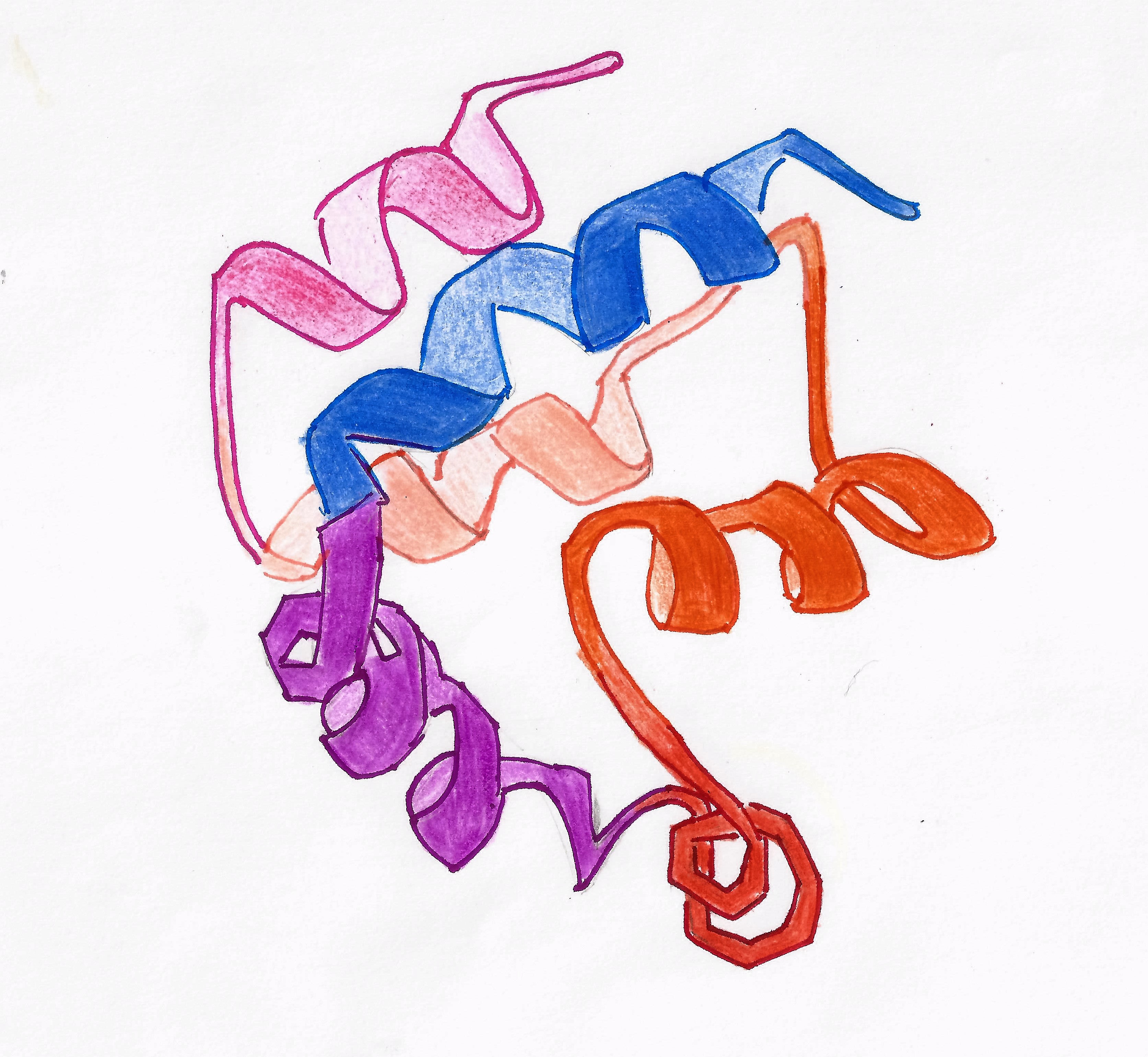
Death Effector Domain
The death-effector domain (DED) is a protein interaction domain found only in eukaryotes that regulates a variety of cellular signalling pathways. The DED domain is found in inactive procaspases (cysteine proteases) and proteins that regulate caspase activation in the apoptosis cascade such as FAS-associating death domain-containing protein (FADD). FADD recruits procaspase 8 and procaspase 10 into a death induced signaling complex (DISC). This recruitment is mediated by a homotypic interaction between the procaspase DED and a second DED that is death effector domain in an adaptor protein that is directly associated with activated TNF receptors. Complex formation allows proteolytic activation of procaspase into the active caspase form which results in the initiation of apoptosis (cell death). Structurally the DED domain are a subclass of protein motif known as the death fold and contains 6 alpha helices, that closely resemble the structure of the Death domain (DD). Structure DE ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FADD
FAS-associated death domain protein, also called MORT1, is encoded by the ''FADD'' gene on the 11q13.3 region of chromosome 11 in humans. FADD is an adaptor protein that bridges members of the tumor necrosis factor receptor superfamily, such as the Fas-receptor, to procaspases 8 and 10 to form the death-inducing signaling complex (DISC) during apoptosis. As well as its most well known role in apoptosis, FADD has also been seen to play a role in other processes including proliferation, cell cycle regulation and development. Structure FADD is a 23 kDa protein, made up of 208 amino acids. It contains two main domains: a C terminal death domain (DD) and an N terminal death effector domain (DED). Each domain, although sharing very little sequence similarity, are structurally similar to one another, with each consisting of 6 α helices. The DD of FADD binds to receptors such as the Fas receptor at the plasma membrane via their DD. The interaction between the death domains are elec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |