|
Biomarker Discovery
Biomarker discovery is a medical term describing the process by which biomarkers are discovered. Many commonly used blood tests in medicine are biomarkers. There is interest in biomarker discovery on the part of the pharmaceutical industry; blood-test or other biomarkers could serve as intermediate markers of disease in clinical trials, and as possible drug targets. Mechanism of action The way that these tests have been found can be viewed as biomarker discovery; however, their identification has primarily been made one at a time. Many well-known tests have been identified based on biological insight from the fields of physiology or biochemistry; therefore, only a few markers at a time have been considered. An example of biomarker discovery is the use of inulin to assess kidney function. From this process a naturally occurring molecule (creatinine) was discovered, enabling the same measurements to be made without insulin injections. The recent interest in biomarker discovery is spu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biomarker (medicine)
In medicine, a biomarker is a measurable indicator of the severity or presence of some disease state. More generally a biomarker is anything that can be used as an indicator of a particular disease state or some other physiological state of an organism. According to the WHO, the indicator may be chemical, physical, or biological in nature - and the measurement may be functional, physiological, biochemical, cellular, or molecular. A biomarker can be a substance that is introduced into an organism as a means to examine organ function or other aspects of health. For example, rubidium chloride is used in isotopic labeling to evaluate perfusion of heart muscle. It can also be a substance whose detection indicates a particular disease state, for example, the presence of an antibody may indicate an infection. More specifically, a biomarker indicates a change in expression or state of a protein that correlates with the risk or progression of a disease, or with the susceptibility of the dis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-reactive Protein
C-reactive protein (CRP) is an annular (ring-shaped) pentameric protein found in blood plasma, whose circulating concentrations rise in response to inflammation. It is an acute-phase protein of hepatic origin that increases following interleukin-6 secretion by macrophages and T cells. Its physiological role is to bind to lysophosphatidylcholine expressed on the surface of dead or dying cells (and some types of bacteria) in order to activate the complement system via C1q. CRP is synthesized by the liver in response to factors released by macrophages and fat cells (adipocytes). It is a member of the pentraxin family of proteins. It is not related to C-peptide (insulin) or protein C (blood coagulation). C-reactive protein was the first pattern recognition receptor (PRR) to be identified. History Discovered by Tillett and Francis in 1930, it was initially thought that CRP might be a pathogenic secretion since it was elevated in a variety of illnesses, including cancer. The later ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tissue Microarray
Tissue microarrays (also TMAs) consist of paraffin blocks in which up to 1000 separate tissue cores are assembled in array fashion to allow multiplex histological analysis. History The major limitations in molecular clinical analysis of tissues include the cumbersome nature of procedures, limited availability of diagnostic reagents and limited patient sample size. The technique of tissue microarray was developed to address these issues. Multi-tissue blocks were first introduced by H. Battifora in 1986 with his so-called “multitumor (sausage) tissue block" and modified in 1990 with its improvement, "the checkerboard tissue block" . In 1998, J. Kononen and collaborators developed the current technique, which uses a novel sampling approach to produce tissues of regular size and shape that can be more densely and precisely arrayed. Procedure In the tissue microarray technique, a hollow needle is used to remove tissue cores as small as 0.6 mm in diameter from regions of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antibody Microarray
An antibody microarray (also known as antibody array) is a specific form of protein microarray. In this technology, a collection of captured antibodies are spotted and fixed on a solid surface such as glass, plastic, membrane, or silicon chip, and the interaction between the antibody and its target antigen is detected. Antibody microarrays are often used for detecting protein expression from various biofluids including serum, plasma and cell or tissue lysates. Antibody arrays may be used for both basic research and medical and diagnostic applications. Background The concept and methodology of antibody microarrays were first introduced by Tse Wen Chang in 1983 in a scientific publication and a series of patents, when he was working at Centocor in Malvern, Pennsylvania. Chang coined the term “antibody matrix” and discussed “array” arrangement of minute antibody spots on small glass or plastic surfaces. He demonstrated that a 10×10 (100 in total) and 20×20 (400 in total) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Matrix-assisted Laser Desorption/ionization
In mass spectrometry, matrix-assisted laser desorption/ionization (MALDI) is an ionization technique that uses a laser energy absorbing matrix to create ions from large molecules with minimal fragmentation. It has been applied to the analysis of biomolecules ( biopolymers such as DNA, proteins, peptides and carbohydrates) and various organic molecules (such as polymers, dendrimers and other macromolecules), which tend to be fragile and fragment when ionized by more conventional ionization methods. It is similar in character to electrospray ionization (ESI) in that both techniques are relatively soft (low fragmentation) ways of obtaining ions of large molecules in the gas phase, though MALDI typically produces far fewer multi-charged ions. MALDI methodology is a three-step process. First, the sample is mixed with a suitable matrix material and applied to a metal plate. Second, a pulsed laser irradiates the sample, triggering ablation and desorption of the sample and matrix materi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Surface-enhanced Laser Desorption/ionization
Surface-enhanced laser desorption/ionization (SELDI) is a soft ionization method in mass spectrometry (MS) used for the analysis of protein mixtures. It is a variation of matrix-assisted laser desorption/ionization (MALDI). In MALDI, the sample is mixed with a matrix material and applied to a metal plate before irradiation by a laser, whereas in SELDI, proteins of interest in a sample become bound to a surface before MS analysis. The sample surface is a key component in the purification, desorption, and ionization of the sample. SELDI is typically used with time-of-flight (TOF) mass spectrometers and is used to detect proteins in tissue samples, blood, urine, or other clinical samples, however, SELDI technology can potentially be used in any application by simply modifying the sample surface. Sample preparation and instrumentation SELDI can be seen as a combination of solid-phase chromatography and TOF-MS. The sample is applied to a modified chip surface, which allows for the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
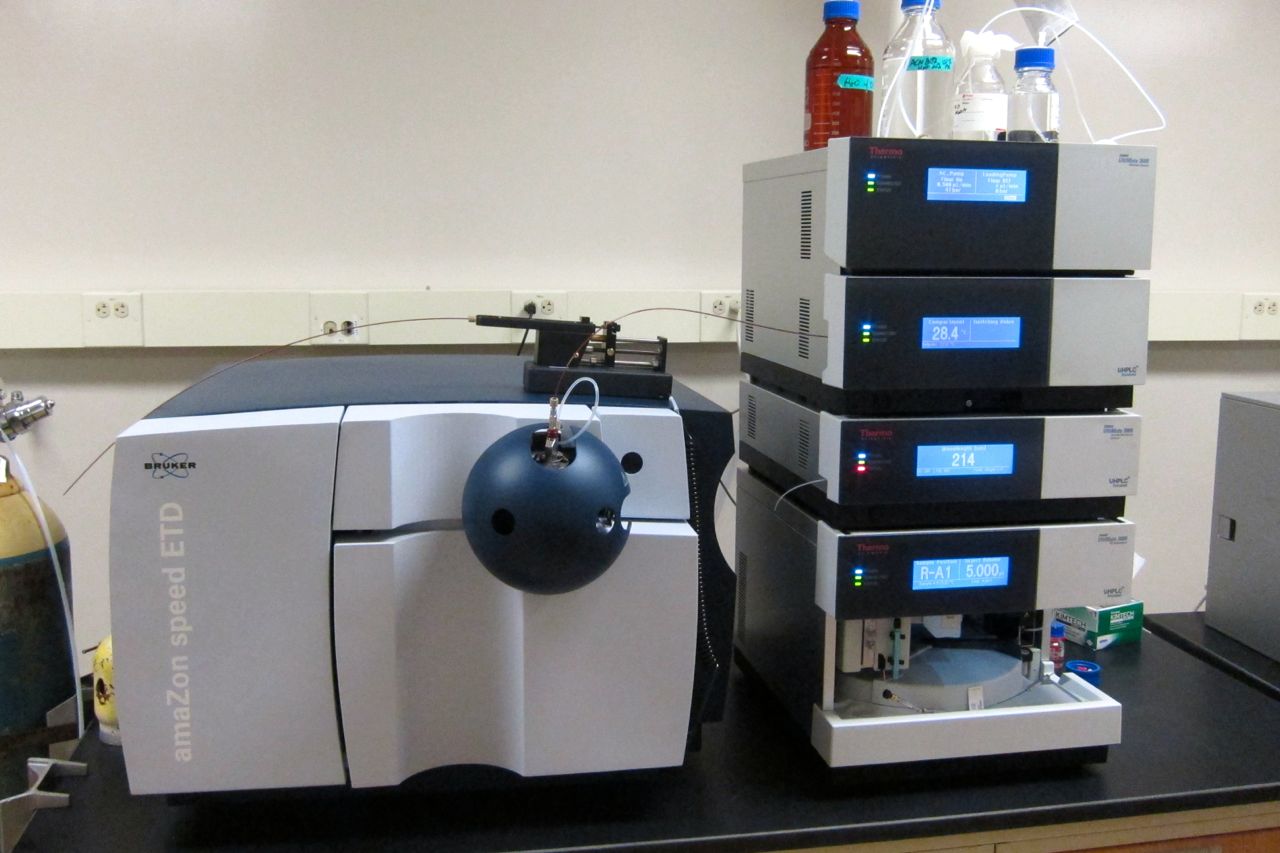
Liquid Chromatography–mass Spectrometry
Liquid chromatography–mass spectrometry (LC–MS) is an analytical chemistry technique that combines the physical separation capabilities of liquid chromatography (or HPLC) with the mass analysis capabilities of mass spectrometry (MS). Coupled chromatography - MS systems are popular in chemical analysis because the individual capabilities of each technique are enhanced synergistically. While liquid chromatography separates mixtures with multiple components, mass spectrometry provides spectral information that may help to identify (or confirm the suspected identity of) each separated component. MS is not only sensitive, but provides selective detection, relieving the need for complete chromatographic separation. LC-MS is also appropriate for metabolomics because of its good coverage of a wide range of chemicals. This tandem technique can be used to analyze biochemical, organic, and inorganic compounds commonly found in complex samples of environmental and biological origin. There ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Two-dimensional Gel Electrophoresis
Two-dimensional gel electrophoresis, abbreviated as 2-DE or 2-D electrophoresis, is a form of gel electrophoresis commonly used to analyze proteins. Mixtures of proteins are separated by two properties in two dimensions on 2D gels. 2-DE was first independently introduced by O'Farrell and Klose in 1975. Basis for separation 2-D electrophoresis begins with electrophoresis in the first dimension and then separates the molecules perpendicularly from the first to create an electropherogram in the second dimension. In electrophoresis in the first dimension, molecules are separated linearly according to their isoelectric point. In the second dimension, the molecules are then separated at 90 degrees from the first electropherogram according to molecular mass. Since it is unlikely that two molecules will be similar in two distinct properties, molecules are more effectively separated in 2-D electrophoresis than in 1-D electrophoresis. The two dimensions that proteins are separated into ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Microarray
A DNA microarray (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles (10−12 moles) of a specific DNA sequence, known as '' probes'' (or ''reporters'' or '' oligos''). These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA (also called anti-sense RNA) sample (called ''target'') under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was inv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serial Analysis Of Gene Expression
Serial Analysis of Gene Expression (SAGE) is a transcriptomic technique used by molecular biologists to produce a snapshot of the messenger RNA population in a sample of interest in the form of small tags that correspond to fragments of those transcripts. Several variants have been developed since, most notably a more robust version, LongSAGE, RL-SAGE and the most recent SuperSAGE. Many of these have improved the technique with the capture of longer tags, enabling more confident identification of a source gene. Overview Briefly, SAGE experiments proceed as follows: # The mRNA of an input sample (e.g. a tumour) is isolated and a reverse transcriptase and biotinylated primers are used to synthesize cDNA from mRNA. # The cDNA is bound to Streptavidin beads via interaction with the biotin attached to the primers, and is then cleaved using a restriction endonuclease called an anchoring enzyme (AE). The location of the cleavage site and thus the length of the remaining cDNA bound ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |