|
ViennaRNA Package
The ViennaRNA Package is a set of standalone programs and libraries used for prediction and analysis of RNA secondary structures. The source code for the package is distributed freely and compiled binaries are available for Linux, macOS and Windows platforms. The original paper has been cited over 2000 times. Background The three dimensional structure of biological macromolecules like proteins and nucleic acids play critical role in determining their functional role. This process of decoding function from the sequence is an experimentally and computationally challenging question addressed widely. RNA structures form complex secondary and tertiary structures compared to DNA which form duplexes with full complementarity between two strands. This is partially because the extra oxygen in RNA increases the propensity for hydrogen bonding in the nucleic acid backbone. The base pairing and base stacking interactions of RNA play critical role in formation of ribosome, spliceosome, o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C (programming Language)
C (''pronounced like the letter c'') is a General-purpose language, general-purpose computer programming language. It was created in the 1970s by Dennis Ritchie, and remains very widely used and influential. By design, C's features cleanly reflect the capabilities of the targeted CPUs. It has found lasting use in operating systems, device drivers, protocol stacks, though decreasingly for application software. C is commonly used on computer architectures that range from the largest supercomputers to the smallest microcontrollers and embedded systems. A successor to the programming language B (programming language), B, C was originally developed at Bell Labs by Ritchie between 1972 and 1973 to construct utilities running on Unix. It was applied to re-implementing the kernel of the Unix operating system. During the 1980s, C gradually gained popularity. It has become one of the measuring programming language popularity, most widely used programming languages, with C compilers avail ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Base Stacking
Nucleic acid tertiary structure is the three-dimensional shape of a nucleic acid polymer. RNA and DNA molecules are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three-dimensional structure. While such structures are diverse and seemingly complex, they are composed of recurring, easily recognizable tertiary structural motifs that serve as molecular building blocks. Some of the most common motifs for RNA and DNA tertiary structure are described below, but this information is based on a limited number of solved structures. Many more tertiary structural motifs will be revealed as new RNA and DNA molecules are structurally characterized. Helical structures Double helix The double helix is the dominant tertiary structure for biological DNA, and is also a possible structure for RNA. Three DNA conformations are believed to be found in nature, A-DNA, B-DNA, and Z-DNA. The "B" form described by James D. Wats ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics Software
The list of bioinformatics software tools can be split up according to the license used: * List of proprietary bioinformatics software *List of open-source bioinformatics software Alternatively, here is a categorization according to the respective bioinformatics subfield specialized on: *Sequence analysis software **List of sequence alignment software **List of alignment visualization software **Alignment-free sequence analysis **De novo sequence assemblers **List of gene prediction software ** List of disorder prediction software ** List of Protein subcellular localization prediction tools ** List of phylogenetics software ** List of phylogenetic tree visualization software ** :Metagenomics_software *Structural biology software **List of molecular graphics systems **List of protein-ligand docking software ** List of RNA structure prediction software ** List of software for protein model error verification **List of protein secondary structure prediction programs **List of protein st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Structure Prediction
Nucleic acid structure prediction is a computational method to determine ''secondary'' and ''tertiary'' nucleic acid structure from its sequence. Secondary structure can be predicted from one or several nucleic acid sequences. Tertiary structure can be predicted from the sequence, or by comparative modeling (when the structure of a homologous sequence is known). The problem of predicting nucleic acid secondary structure is dependent mainly on base pairing and base stacking interactions; many molecules have several possible three-dimensional structures, so predicting these structures remains out of reach unless obvious sequence and functional similarity to a known class of nucleic acid molecules, such as transfer RNA (tRNA) or microRNA (miRNA), is observed. Many secondary structure prediction methods rely on variations of dynamic programming and therefore are unable to efficiently identify pseudoknots. While the methods are similar, there are slight differences in the approache ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Structure Determination
Experimental approaches of determining the structure of nucleic acids, such as RNA and DNA, can be largely classified into biophysical and biochemical methods. Biophysical methods use the fundamental physical properties of molecules for structure determination, including X-ray crystallography, NMR and cryo-EM. Biochemical methods exploit the chemical properties of nucleic acids using specific reagents and conditions to assay the structure of nucleic acids. Such methods may involve chemical probing with specific reagents, or rely on native or analogue chemistry. Different experimental approaches have unique merits and are suitable for different experimental purposes. Biophysical methods X-ray crystallography X-ray crystallography is not common for nucleic acids alone, since neither DNA nor RNA readily form crystals. This is due to the greater degree of intrinsic disorder and dynamism in nucleic acid structures and the negatively charged (deoxy)ribose-phosphate backbones, whic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics. History The original FASTA program was designed for protein sequence similarity searching. Because of the exponentially expanding genetic information and the limited speed and memory of computers in the 1980s heuristic methods were introduced aligning a query sequence to entire data-bases. FASTA, published in 1987, added the ability to do DNA:DNA searches, translated protein:DNA searches, and also provided a more sophisticated shuffling program for evaluating statistical significance. There are several programs in this package that allow the alignment of protein sequences and DNA sequences. Nowadays, increased computer performance makes it possible to perform searches for local alignment detection in a database using the Smith–Waterman algorithm. FASTA is pronounced "f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Application Programming Interface
An application programming interface (API) is a way for two or more computer programs to communicate with each other. It is a type of software interface, offering a service to other pieces of software. A document or standard that describes how to build or use such a connection or interface is called an ''API specification''. A computer system that meets this standard is said to ''implement'' or ''expose'' an API. The term API may refer either to the specification or to the implementation. In contrast to a user interface, which connects a computer to a person, an application programming interface connects computers or pieces of software to each other. It is not intended to be used directly by a person (the end user) other than a computer programmer who is incorporating it into the software. An API is often made up of different parts which act as tools or services that are available to the programmer. A program or a programmer that uses one of these parts is said to ''call'' tha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cluster Analysis
Cluster analysis or clustering is the task of grouping a set of objects in such a way that objects in the same group (called a cluster) are more similar (in some sense) to each other than to those in other groups (clusters). It is a main task of exploratory data analysis, and a common technique for statistical data analysis, used in many fields, including pattern recognition, image analysis, information retrieval, bioinformatics, data compression, computer graphics and machine learning. Cluster analysis itself is not one specific algorithm, but the general task to be solved. It can be achieved by various algorithms that differ significantly in their understanding of what constitutes a cluster and how to efficiently find them. Popular notions of clusters include groups with small distances between cluster members, dense areas of the data space, intervals or particular statistical distributions. Clustering can therefore be formulated as a multi-objective optimization probl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dynamic Programming
Dynamic programming is both a mathematical optimization method and a computer programming method. The method was developed by Richard Bellman in the 1950s and has found applications in numerous fields, from aerospace engineering to economics. In both contexts it refers to simplifying a complicated problem by breaking it down into simpler sub-problems in a recursive manner. While some decision problems cannot be taken apart this way, decisions that span several points in time do often break apart recursively. Likewise, in computer science, if a problem can be solved optimally by breaking it into sub-problems and then recursively finding the optimal solutions to the sub-problems, then it is said to have '' optimal substructure''. If sub-problems can be nested recursively inside larger problems, so that dynamic programming methods are applicable, then there is a relation between the value of the larger problem and the values of the sub-problems.Cormen, T. H.; Leiserson, C. E.; Riv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
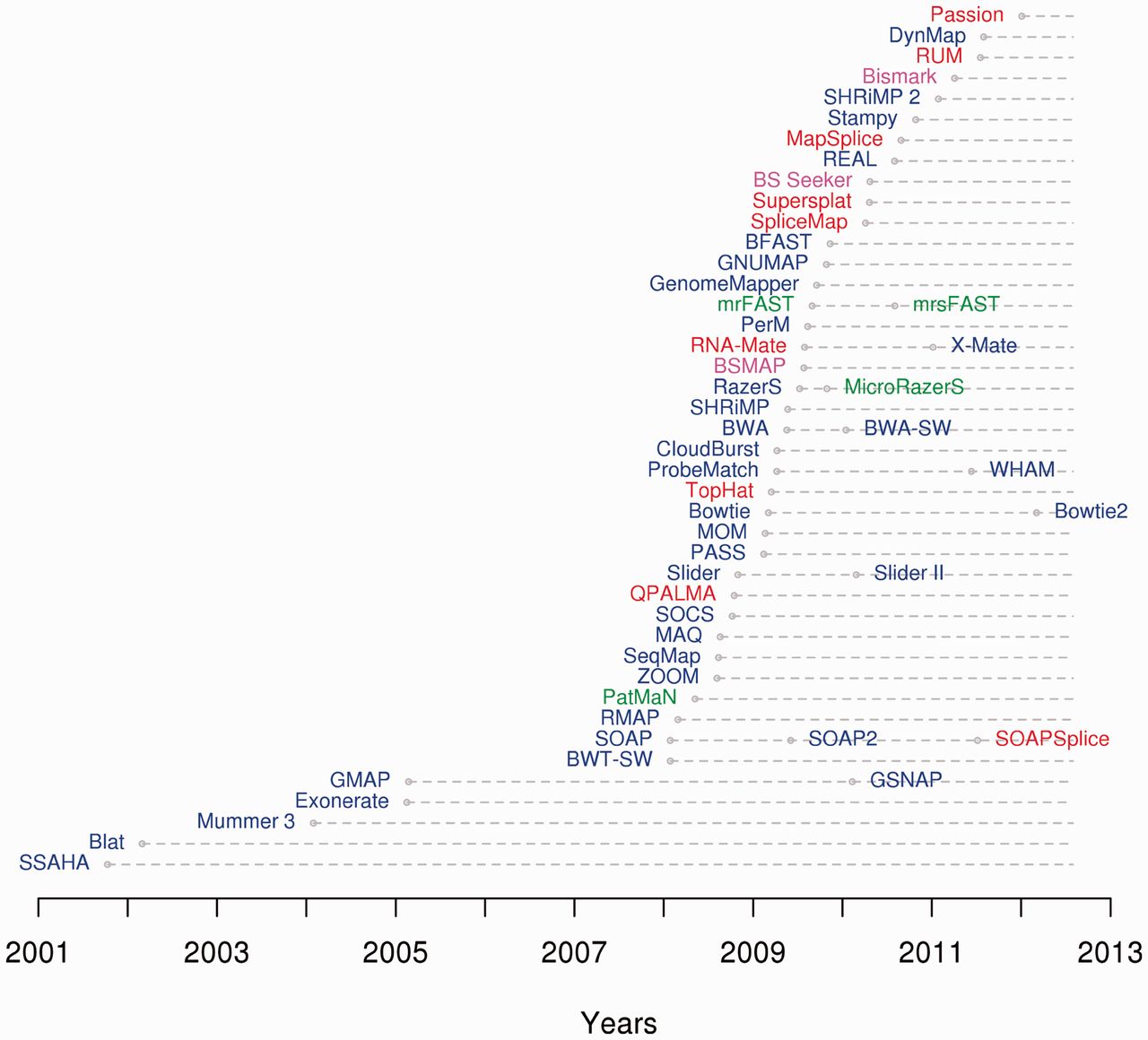
List Of RNA Structure Prediction Software
This list of RNA structure prediction software is a compilation of software tools and web portals used for RNA structure prediction. Single sequence secondary structure prediction. Single sequence tertiary structure prediction Comparative methods The single sequence methods mentioned above have a difficult job detecting a small sample of reasonable secondary structures from a large space of possible structures. A good way to reduce the size of the space is to use evolutionary approaches. Structures that have been conserved by evolution are far more likely to be the functional form. The methods below use this approach. RNA solvent accessibility prediction Intermolecular interactions: RNA-RNA Many ncRNAs function by binding to other RNAs. For example, miRNAs regulate protein coding gene expression by binding to 3' UTRs, small nucleolar RNAs guide post-transcriptional modifications by binding to rRNA, U4 spliceosomal RNA and U6 spliceosomal RNA bind to each other forming pa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Structure
Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA. Chemically speaking, DNA and RNA are very similar. Nucleic acid structure is often divided into four different levels: primary, secondary, tertiary, and quaternary. Primary structure Primary structure consists of a linear sequence of nucleotides that are linked together by phosphodiester bond. It is this linear sequence of nucleotides that make up the primary structure of DNA or RNA. Nucleotides consist of 3 components: # Nitrogenous base ## Adenine ## Guanine ## Cytosine ## Thymine (present in DNA only) ## Uracil (present in RNA only) # 5-carbon sugar which is called deoxyribose (found in DNA) and ribose (found in RNA). # One or more phosphate groups. The nitrogen bases adenine and guanine are purine in structure and form a glycosidic bond between their 9 nitrogen and the 1' -OH group of the deoxyribose. Cytosine, thymine, and uracil are pyrimidines, hence the glycosidic bonds form betwe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TRNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino acid sequence of proteins. tRNAs genes from Bacteria are typically shorter (mean = 77.6 bp) than tRNAs from Archaea (mean = 83.1 bp) and eukaryotes (mean = 84.7 bp). The mature tRNA follows an opposite pattern with tRNAs from Bacteria being usually longer (median = 77.6 nt) than tRNAs from Archaea (median = 76.8 nt), with eukaryotes exhibiting the shortest mature tRNAs (median = 74.5 nt). Transfer RNA (tRNA) does this by carrying an amino acid to the protein synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |