|
Unsolved Problems In Biology
This article lists notable unsolved problems in biology. General biology Evolution and origins of life *Origin of life. Exactly how, where, and when did life on Earth originate? Which, if any, of the many hypotheses is correct? What were the metabolic pathways used by the earliest life forms? * Origins of viruses. Exactly how and when did different groups of viruses originate? *Development and evolution of brain. How and why did the brain evolve? What are the molecular determinants of individual brain development? *Origin of Eukaryotes (Symbiogenesis). How and why did cells combine to form the eukaryotic cell? Did one or more random events lead to the first eukaryotic cells, or can the formation of eukaryotic cells be explained by physical and biological principles? How did the mitochondria's mitosis cycle come in sync with its host cell? Did the mitochondria or the nucleus develop first in eukaryotes? *Last Universal Common Ancestor. What were the characteristics of the Last U ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Unsolved Problems
List of unsolved problems may refer to several notable conjectures or open problems in various academic fields: Natural sciences, engineering and medicine * Unsolved problems in astronomy * Unsolved problems in biology * Unsolved problems in chemistry * Unsolved problems in engineering * Unsolved problems in geoscience * Unsolved problems in medicine * Unsolved problems in neuroscience * Unsolved problems in physics Mathematics, philosophy and information sciences * Unsolved problems in mathematics * Unsolved problems in statistics * Unsolved problems in computer science * Unsolved problems in philosophy * Unsolved problems in information theory Social sciences and languages * Unsolved problems in economics * Unsolved problems in fair division * Unsolved problems in linguistics See also * Cold case (unsolved crimes) * List of ciphertexts * List of NP-complete problems * List of PSPACE-complete problems * List of paradoxes * List of undecidable problems * List of unsolv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ketamine
Ketamine is a dissociative anesthetic used medically for induction and maintenance of anesthesia. It is also used as a recreational drug. It is one of the safest anesthetics, as, in contrast with opiates, ether, and propofol, it suppresses neither respiration nor heart rate. Ketamine is also simple to administer and highly tolerable compared to drugs with similar effects which are flammable, irritating, or even explosive. Ketamine is a novel compound, derived from PCP, created in pursuit of a safer anesthetic with similar characteristics. Ketamine is also used for acute pain management. At anesthetic doses, ketamine induces a state of "dissociative anesthesia", a trance-like state providing pain relief, sedation, and amnesia. The distinguishing features of ketamine anesthesia are preserved breathing and airway reflexes, stimulated heart function with increased blood pressure, and moderate bronchodilation. At lower, sub-anesthetic doses, ketamine is a promising agent for pain ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Orphan Receptor
In biochemistry, an orphan receptor is a protein that has a similar structure to other identified receptors but whose endogenous ligand has not yet been identified. If a ligand for an orphan receptor is later discovered, the receptor is referred to as an "adopted orphan". Conversely, the term orphan ligand refers to a biological ligand whose cognate receptor has not yet been identified. Examples Examples of orphan receptors are found in the G protein-coupled receptor (GPCR) and nuclear receptor families. If an endogenous ligand is found, the orphan receptor is "adopted" or "de-orphanized". An example is the nuclear receptor Farnesoid X receptor (FXR) and the GPCR TGR5/GPCR19/G protein-coupled bile acid receptor, both of which are activated by bile acids. Adopted orphan receptors in the nuclear receptor group include FXR, liver X receptor (LXR), and peroxisome proliferator-activated receptor (PPAR). Another example of an orphan receptor site is the PCP binding site in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligand (biochemistry)
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure. Binding occurs by intermolecular forces, such as ionic bonds, hydrogen bonds and Van der Waals forces. The association or docking is actually reversible through dissociation. Measurably irreversible covalent bonding between a ligand and target molecule is atypical in biological systems. In contrast to the definition of lig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequential Model
The sequential model (also known as the KNF model) is a theory that describes cooperativity of protein subunits. Koshland, D.E., Némethy, G. and Filmer, D. (1966) Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5, 365–385DOI: 10.1021/bi00865a047/ref> It postulates that a protein's conformation changes with each binding of a ligand, thus sequentially changing its affinity for the ligand at neighboring binding sites. It gives one explanation for cooperative binding. Overview This model for allosteric regulation of enzymes suggests that the subunits of multimeric proteins have two conformational states. The binding of the ligand causes conformational change in the other subunits of the multimeric protein. Although the subunits go through conformational changes independently (as opposed to in the MWC model), the switch of one subunit makes the other subunits more likely to change, by reducing the energy needed for subseq ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monod-Wyman-Changeux Model
In biochemistry, the Monod-Wyman-Changeux model (MWC model, also known as the symmetry model) describes allosteric transitions of proteins made up of identical subunits. It was proposed by Jean-Pierre Changeux in his PhD thesis, and described by Jacques Monod, Jeffries Wyman, and Jean-Pierre Changeux. It contrasts with the sequential model. The concept of two distinct symmetric states is the central postulate of the MWC model. The main idea is that regulated proteins, such as many enzymes and receptors, exist in different interconvertible states ''in the absence of any regulator''. The ratio of the different conformational states is determined by thermal equilibrium. This model is defined by the following rules: # An allosteric protein is an oligomer of protomers that are symmetrically related (for hemoglobin, we shall assume, for the sake of algebraic simplicity, that all four subunits are functionally identical). # Each protomer can exist in (at least) two conformational sta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
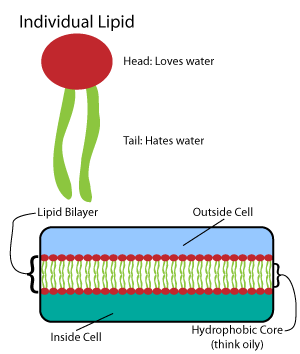
Biosynthesis
Biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism. The prerequisite elements for biosynthesis include: precursor compounds, chemical energy (e.g. ATP), and catalytic enzymes which may require coenzymes (e.g.NADH, NADPH). These elements create monomers, the building blocks for macromolecules. Some important biological macromolecules include: proteins, which are composed of amino acid monomers joined via peptide bon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Design
Protein design is the rational design of new protein molecules to design novel activity, behavior, or purpose, and to advance basic understanding of protein function. Proteins can be designed from scratch (''de novo'' design) or by making calculated variants of a known protein structure and its sequence (termed ''protein redesign''). Rational protein design approaches make protein-sequence predictions that will fold to specific structures. These predicted sequences can then be validated experimentally through methods such as peptide synthesis, site-directed mutagenesis, or artificial gene synthesis. Rational protein design dates back to the mid-1970s. Recently, however, there were numerous examples of successful rational design of water-soluble and even transmembrane peptides and proteins, in part due to a better understanding of different factors contributing to protein structure stability and development of better computational methods. Overview and history The goal in ration ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme Kinetics
Enzyme kinetics is the study of the rates of enzyme-catalysed chemical reactions. In enzyme kinetics, the reaction rate is measured and the effects of varying the conditions of the reaction are investigated. Studying an enzyme's kinetics in this way can reveal the catalytic mechanism of this enzyme, its role in metabolism, how its activity is controlled, and how a drug or a modifier ( inhibitor or activator) might affect the rate. An enzyme (E) is typically a protein molecule that promotes a reaction of another molecule, its substrate (S). This binds to the active site of the enzyme to produce an enzyme-substrate complex ES, and is transformed into an enzyme-product complex EP and from there to product P, via a transition state ES*. The series of steps is known as the mechanism: : E + S ⇄ ES ⇄ ES* ⇄ EP ⇄ E + P This example assumes the simplest case of a reaction with one substrate and one product. Such cases exist: for example, a mutase such as phosphoglucomutase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DeepMind AlphaGo
AlphaGo is a computer program that plays the board game Go. It was developed by DeepMind Technologies a subsidiary of Google (now Alphabet Inc.). Subsequent versions of AlphaGo became increasingly powerful, including a version that competed under the name Master. After retiring from competitive play, AlphaGo Master was succeeded by an even more powerful version known as AlphaGo Zero, which was completely self-taught without learning from human games. AlphaGo Zero was then generalized into a program known as AlphaZero, which played additional games, including chess and shogi. AlphaZero has in turn been succeeded by a program known as MuZero which learns without being taught the rules. AlphaGo and its successors use a Monte Carlo tree search algorithm to find its moves based on knowledge previously acquired by machine learning, specifically by an artificial neural network (a deep learning method) by extensive training, both from human and computer play. A neural network is trai ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
AlphaFold
AlphaFold is an artificial intelligence (AI) program developed by DeepMind, a subsidiary of Alphabet Inc., Alphabet, which performs Protein structure prediction, predictions of protein structure. The program is designed as a deep learning system. AlphaFold AI software has had two major versions. A team of researchers that used AlphaFold 1 (2018) placed first in the overall rankings of the 13th Critical Assessment of protein Structure Prediction (CASP) in December 2018. The program was particularly successful at predicting the most accurate structure for targets rated as the most difficult by the competition organisers, where no existing Threading (protein sequence), template structures were available from proteins with a partially similar sequence. A team that used AlphaFold 2 (2020) repeated the placement in the CASP competition in November 2020. The team achieved a level of accuracy much higher than any other group. It scored above 90 for around two-thirds of the proteins in C ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |