Protein design on:
[Wikipedia]
[Google]
[Amazon]
Protein design is the
 Protein function is heavily dependent on protein structure, and rational protein design uses this relationship to design function by designing proteins that have a target structure or fold. Thus, by definition, in rational protein design the target structure or ensemble of structures must be known beforehand. This contrasts with other forms of protein engineering, such as
Protein function is heavily dependent on protein structure, and rational protein design uses this relationship to design function by designing proteins that have a target structure or fold. Thus, by definition, in rational protein design the target structure or ensemble of structures must be known beforehand. This contrasts with other forms of protein engineering, such as
 In rational protein design, proteins can be redesigned from the sequence and structure of a known protein, or completely from scratch in ''de novo'' protein design. In protein redesign, most of the residues in the sequence are maintained as their wild-type amino-acid while a few are allowed to mutate. In ''de novo'' design, the entire sequence is designed anew, based on no prior sequence.
Both ''de novo'' designs and protein redesigns can establish rules on the
In rational protein design, proteins can be redesigned from the sequence and structure of a known protein, or completely from scratch in ''de novo'' protein design. In protein redesign, most of the residues in the sequence are maintained as their wild-type amino-acid while a few are allowed to mutate. In ''de novo'' design, the entire sequence is designed anew, based on no prior sequence.
Both ''de novo'' designs and protein redesigns can establish rules on the
 Rational protein design techniques must be able to discriminate sequences that will be stable under the target fold from those that would prefer other low-energy competing states. Thus, protein design requires accurate energy functions that can rank and score sequences by how well they fold to the target structure. At the same time, however, these energy functions must consider the computational challenges behind protein design. One of the most challenging requirements for successful design is an energy function that is both accurate and simple for computational calculations.
The most accurate energy functions are those based on quantum mechanical simulations. However, such simulations are too slow and typically impractical for protein design. Instead, many protein design algorithms use either physics-based energy functions adapted from
Rational protein design techniques must be able to discriminate sequences that will be stable under the target fold from those that would prefer other low-energy competing states. Thus, protein design requires accurate energy functions that can rank and score sequences by how well they fold to the target structure. At the same time, however, these energy functions must consider the computational challenges behind protein design. One of the most challenging requirements for successful design is an energy function that is both accurate and simple for computational calculations.
The most accurate energy functions are those based on quantum mechanical simulations. However, such simulations are too slow and typically impractical for protein design. Instead, many protein design algorithms use either physics-based energy functions adapted from 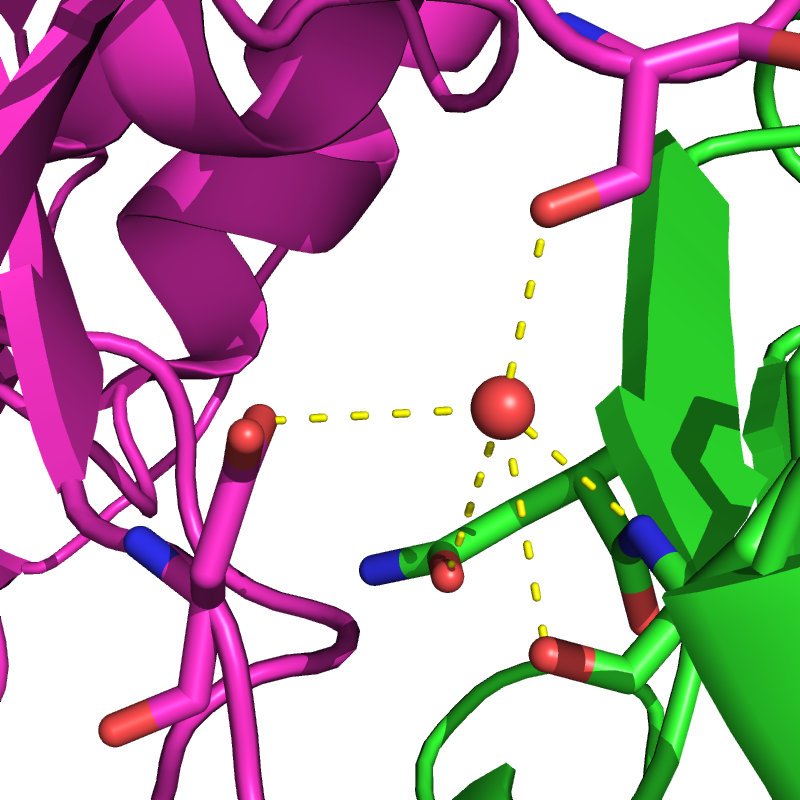 Statistical potentials, in contrast to physics-based potentials, have the advantage of being fast to compute, of accounting implicitly of complex effects and being less sensitive to small changes in the protein structure. These energy functions are based on deriving energy values from frequency of appearance on a structural database.
Protein design, however, has requirements that can sometimes be limited in molecular mechanics force-fields. Molecular mechanics force-fields, which have been
used mostly in molecular dynamics simulations, are optimized for the simulation of single sequences, but protein design searches through many conformations of many sequences. Thus, molecular mechanics force-fields must be tailored for protein design. In practice, protein design energy functions often incorporate both statistical terms and physics-based terms. For example, the Rosetta energy function, one of the most-used energy functions, incorporates physics-based energy terms originating in the CHARMM energy function, and statistical energy terms, such as rotamer probability and knowledge-based electrostatics. Typically, energy functions are highly customized between laboratories, and specifically tailored for every design.
Statistical potentials, in contrast to physics-based potentials, have the advantage of being fast to compute, of accounting implicitly of complex effects and being less sensitive to small changes in the protein structure. These energy functions are based on deriving energy values from frequency of appearance on a structural database.
Protein design, however, has requirements that can sometimes be limited in molecular mechanics force-fields. Molecular mechanics force-fields, which have been
used mostly in molecular dynamics simulations, are optimized for the simulation of single sequences, but protein design searches through many conformations of many sequences. Thus, molecular mechanics force-fields must be tailored for protein design. In practice, protein design energy functions often incorporate both statistical terms and physics-based terms. For example, the Rosetta energy function, one of the most-used energy functions, incorporates physics-based energy terms originating in the CHARMM energy function, and statistical energy terms, such as rotamer probability and knowledge-based electrostatics. Typically, energy functions are highly customized between laboratories, and specifically tailored for every design.
 The goal of protein design is to find a protein sequence that will fold to a target structure. A protein design algorithm must, thus, search all the conformations of each sequence, with respect to the target fold, and rank sequences according to the lowest-energy conformation of each one, as determined by the protein design energy function. Thus, a typical input to the protein design algorithm is the target fold, the sequence space, the structural flexibility, and the energy function, while the output is one or more sequences that are predicted to fold stably to the target structure.
The number of candidate protein sequences, however, grows exponentially with the number of protein residues; for example, there are 20100 protein sequences of length 100. Furthermore, even if amino acid side-chain conformations are limited to a few rotamers (see Structural flexibility), this results in an exponential number of conformations for each sequence. Thus, in our 100 residue protein, and assuming that each amino acid has exactly 10 rotamers, a search algorithm that searches this space will have to search over 200100 protein conformations.
The most common energy functions can be decomposed into pairwise terms between rotamers and amino acid types, which casts the problem as a combinatorial one, and powerful optimization algorithms can be used to solve it. In those cases, the total energy of each conformation belonging to each sequence can be formulated as a sum of individual and pairwise terms between residue positions. If a designer is interested only in the best sequence, the protein design algorithm only requires the lowest-energy conformation of the lowest-energy sequence. In these cases, the amino acid identity of each rotamer can be ignored and all rotamers belonging to different amino acids can be treated the same. Let ri be a rotamer at residue position i in the protein chain, and E(ri) the potential energy between the internal atoms of the rotamer. Let E(ri, rj) be the potential energy between ri and rotamer rj at residue position j. Then, we define the optimization problem as one of finding the conformation of minimum energy (ET):
The problem of minimizing ET is an
The goal of protein design is to find a protein sequence that will fold to a target structure. A protein design algorithm must, thus, search all the conformations of each sequence, with respect to the target fold, and rank sequences according to the lowest-energy conformation of each one, as determined by the protein design energy function. Thus, a typical input to the protein design algorithm is the target fold, the sequence space, the structural flexibility, and the energy function, while the output is one or more sequences that are predicted to fold stably to the target structure.
The number of candidate protein sequences, however, grows exponentially with the number of protein residues; for example, there are 20100 protein sequences of length 100. Furthermore, even if amino acid side-chain conformations are limited to a few rotamers (see Structural flexibility), this results in an exponential number of conformations for each sequence. Thus, in our 100 residue protein, and assuming that each amino acid has exactly 10 rotamers, a search algorithm that searches this space will have to search over 200100 protein conformations.
The most common energy functions can be decomposed into pairwise terms between rotamers and amino acid types, which casts the problem as a combinatorial one, and powerful optimization algorithms can be used to solve it. In those cases, the total energy of each conformation belonging to each sequence can be formulated as a sum of individual and pairwise terms between residue positions. If a designer is interested only in the best sequence, the protein design algorithm only requires the lowest-energy conformation of the lowest-energy sequence. In these cases, the amino acid identity of each rotamer can be ignored and all rotamers belonging to different amino acids can be treated the same. Let ri be a rotamer at residue position i in the protein chain, and E(ri) the potential energy between the internal atoms of the rotamer. Let E(ri, rj) be the potential energy between ri and rotamer rj at residue position j. Then, we define the optimization problem as one of finding the conformation of minimum energy (ET):
The problem of minimizing ET is an
rational design
In chemical biology and biomolecular engineering, rational design (RD) is an umbrella term which invites the strategy of creating new molecules with a certain functionality, based upon the ability to predict how the molecule's structure (specific ...
of new protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
molecules to design novel activity, behavior, or purpose, and to advance basic understanding of protein function. Proteins can be designed from scratch (''de novo'' design) or by making calculated variants of a known protein structure and its sequence (termed ''protein redesign''). Rational protein design approaches make protein-sequence predictions that will fold to specific structures. These predicted sequences can then be validated experimentally through methods such as peptide synthesis
In organic chemistry, peptide synthesis is the production of peptides, compounds where multiple amino acids are linked via amide bonds, also known as peptide bonds. Peptides are chemically synthesized by the condensation reaction of the carboxyl ...
, site-directed mutagenesis
Site-directed mutagenesis is a molecular biology method that is used to make specific and intentional mutating changes to the DNA sequence of a gene and any gene products. Also called site-specific mutagenesis or oligonucleotide-directed mutagenes ...
, or artificial gene synthesis.
Rational protein design dates back to the mid-1970s. Recently, however, there were numerous examples of successful rational design of water-soluble and even transmembrane peptides and proteins, in part due to a better understanding of different factors contributing to protein structure stability and development of better computational methods.
Overview and history
The goal in rational protein design is to predictamino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
sequences
In mathematics, a sequence is an enumerated collection of objects in which repetitions are allowed and order matters. Like a set, it contains members (also called ''elements'', or ''terms''). The number of elements (possibly infinite) is call ...
that will fold to a specific protein structure. Although the number of possible protein sequences is vast, growing exponentially with the size of the protein chain, only a subset of them will fold reliably and quickly to one native state
In biochemistry, the native state of a protein or nucleic acid is its properly Protein folding, folded and/or assembled form, which is operative and functional. The native state of a biomolecule may possess all four levels of biomolecular structu ...
. Protein design involves identifying novel sequences within this subset. The native state of a protein is the conformational free energy minimum for the chain. Thus, protein design is the search for sequences that have the chosen structure as a free energy minimum. In a sense, it is the reverse of protein structure prediction
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its Protein secondary structure, secondary and Protein tertiary structure, tertiary structure ...
. In design, a tertiary structure
Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the ...
is specified, and a sequence that will fold to it is identified. Hence, it is also termed ''inverse folding''. Protein design is then an optimization problem: using some scoring criteria, an optimized sequence that will fold to the desired structure is chosen.
When the first proteins were rationally designed during the 1970s and 1980s, the sequence for these was optimized manually based on analyses of other known proteins, the sequence composition, amino acid charges, and the geometry of the desired structure. The first designed proteins are attributed to Bernd Gutte, who designed a reduced version of a known catalyst, bovine ribonuclease, and tertiary structures consisting of beta-sheets and alpha-helices, including a binder of DDT. Urry and colleagues later designed elastin
Elastin is a protein encoded by the ''ELN'' gene in humans and several other animals. Elastin is a key component in the extracellular matrix of gnathostomes (jawed vertebrates). It is highly Elasticity (physics), elastic and present in connective ...
-like fibrous
Fiber (spelled fibre in British English; from ) is a natural or artificial substance that is significantly longer than it is wide. Fibers are often used in the manufacture of other materials. The strongest engineering materials often inco ...
peptides based on rules on sequence composition. Richardson and coworkers designed a 79-residue protein with no sequence homology to a known protein. In the 1990s, the advent of powerful computers, libraries of amino acid conformations, and force fields developed mainly for molecular dynamics
Molecular dynamics (MD) is a computer simulation method for analyzing the Motion (physics), physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamics ( ...
simulations enabled the development of structure-based computational protein design tools. Following the development of these computational tools, great success has been achieved over the last 30 years in protein design. The first protein successfully designed completely ''de novo'' was done by Stephen Mayo and coworkers in 1997, and, shortly after, in 1999 Peter S. Kim and coworkers designed dimers, trimers, and tetramers of unnatural right-handed coiled coil
A coiled coil is a structural motif in proteins in which two to seven alpha-helices are coiled together like the strands of a rope. ( Dimers and trimers are the most common types.) They have been found in roughly 5-10% of proteins and have a ...
s. In 2003, David Baker's laboratory designed a full protein to a fold never seen before in nature. Later, in 2008, Baker's group computationally designed enzymes for two different reactions. In 2010, one of the most powerful broadly neutralizing antibodies was isolated from patient serum using a computationally designed protein probe. In 2024, Baker received one half of the Nobel Prize in Chemistry
The Nobel Prize in Chemistry () is awarded annually by the Royal Swedish Academy of Sciences to scientists in the various fields of chemistry. It is one of the five Nobel Prizes established by the will of Alfred Nobel in 1895, awarded for outst ...
for his advancement of computational protein design, with the other half being shared by Demis Hassabis and John Jumper of Deepmind
DeepMind Technologies Limited, trading as Google DeepMind or simply DeepMind, is a British–American artificial intelligence research laboratory which serves as a subsidiary of Alphabet Inc. Founded in the UK in 2010, it was acquired by Go ...
for protein structure prediction. Due to these and other successes (e.g., see examples
Example may refer to:
* ''exempli gratia'' (e.g.), usually read out in English as "for example"
* .example, reserved as a domain name that may not be installed as a top-level domain of the Internet
** example.com, example.net, example.org, a ...
below), protein design has become one of the most important tools available for protein engineering. There is great hope that the design of new proteins, small and large, will have uses in biomedicine
Biomedicine (also referred to as Western medicine, mainstream medicine or conventional medicine)
and bioengineering
Biological engineering or
bioengineering is the application of principles of biology and the tools of engineering to create usable, tangible, economically viable products. Biological engineering employs knowledge and expertise from a number ...
.
Underlying models of protein structure and function
Protein design programs usecomputer models
Computer simulation is the running of a mathematical model on a computer, the model being designed to represent the behaviour of, or the outcome of, a real-world or physical system. The reliability of some mathematical models can be determin ...
of the molecular forces that drive proteins in ''in vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, an ...
'' environments. In order to make the problem tractable, these forces are simplified by protein design models. Although protein design programs vary greatly, they have to address four main modeling questions: What is the target structure of the design, what flexibility is allowed on the target structure, which sequences are included in the search, and which force field will be used to score sequences and structures.
Target structure
 Protein function is heavily dependent on protein structure, and rational protein design uses this relationship to design function by designing proteins that have a target structure or fold. Thus, by definition, in rational protein design the target structure or ensemble of structures must be known beforehand. This contrasts with other forms of protein engineering, such as
Protein function is heavily dependent on protein structure, and rational protein design uses this relationship to design function by designing proteins that have a target structure or fold. Thus, by definition, in rational protein design the target structure or ensemble of structures must be known beforehand. This contrasts with other forms of protein engineering, such as directed evolution
Directed evolution (DE) is a method used in protein engineering that mimics the process of natural selection to steer proteins or nucleic acids toward a user-defined goal. It consists of subjecting a gene to iterative rounds of mutagenesis (cre ...
, where a variety of methods are used to find proteins that achieve a specific function, and with protein structure prediction
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its Protein secondary structure, secondary and Protein tertiary structure, tertiary structure ...
where the sequence is known, but the structure is unknown.
Most often, the target structure is based on a known structure of another protein. However, novel folds not seen in nature have been made increasingly possible. Peter S. Kim and coworkers designed trimers and tetramers of unnatural coiled coils, which had not been seen before in nature. The protein Top7, developed in David Baker's lab, was designed completely using protein design algorithms, to a completely novel fold. More recently, Baker and coworkers developed a series of principles to design ideal globular-protein structures based on protein folding funnels that bridge between secondary structure prediction and tertiary structures. These principles, which build on both protein structure prediction and protein design, were used to design five different novel protein topologies.
Sequence space
 In rational protein design, proteins can be redesigned from the sequence and structure of a known protein, or completely from scratch in ''de novo'' protein design. In protein redesign, most of the residues in the sequence are maintained as their wild-type amino-acid while a few are allowed to mutate. In ''de novo'' design, the entire sequence is designed anew, based on no prior sequence.
Both ''de novo'' designs and protein redesigns can establish rules on the
In rational protein design, proteins can be redesigned from the sequence and structure of a known protein, or completely from scratch in ''de novo'' protein design. In protein redesign, most of the residues in the sequence are maintained as their wild-type amino-acid while a few are allowed to mutate. In ''de novo'' design, the entire sequence is designed anew, based on no prior sequence.
Both ''de novo'' designs and protein redesigns can establish rules on the sequence space
In functional analysis and related areas of mathematics, a sequence space is a vector space whose elements are infinite sequences of real or complex numbers. Equivalently, it is a function space whose elements are functions from the natural num ...
: the specific amino acids that are allowed at each mutable residue position. For example, the composition of the surface of the RSC3 probe to select HIV-broadly neutralizing antibodies was restricted based on evolutionary data and charge balancing. Many of the earliest attempts on protein design were heavily based on empiric ''rules'' on the sequence space. Moreover, the design of fibrous proteins usually follows strict rules on the sequence space. Collagen
Collagen () is the main structural protein in the extracellular matrix of the connective tissues of many animals. It is the most abundant protein in mammals, making up 25% to 35% of protein content. Amino acids are bound together to form a trip ...
-based designed proteins, for example, are often composed of Gly-Pro-X repeating patterns. The advent of computational techniques allows designing proteins with no human intervention in sequence selection.
Structural flexibility
In protein design, the target structure (or structures) of the protein are known. However, a rational protein design approach must model some ''flexibility'' on the target structure in order to increase the number of sequences that can be designed for that structure and to minimize the chance of a sequence folding to a different structure. For example, in a protein redesign of one small amino acid (such as alanine) in the tightly packed core of a protein, very few mutants would be predicted by a rational design approach to fold to the target structure, if the surrounding side-chains are not allowed to be repacked. Thus, an essential parameter of any design process is the amount of flexibility allowed for both the side-chains and the backbone. In the simplest models, the protein backbone is kept rigid while some of the protein side-chains are allowed to change conformations. However, side-chains can have many degrees of freedom in their bond lengths, bond angles, and χ dihedral angles. To simplify this space, protein design methods use rotamer libraries that assume ideal values for bond lengths and bond angles, while restricting χ dihedral angles to a few frequently observed low-energy conformations termed rotamers. Rotamer libraries are derived from the statistical analysis of many protein structures. Backbone-independent rotamer libraries describe all rotamers. Backbone-dependent rotamer libraries, in contrast, describe the rotamers as how likely they are to appear depending on the protein backbone arrangement around the side chain. Most protein design programs use one conformation (e.g., the modal value for rotamer dihedrals in space) or several points in the region described by the rotamer; the OSPREY protein design program, in contrast, models the entire continuous region. Although rational protein design must preserve the general backbone fold a protein, allowing some backbone flexibility can significantly increase the number of sequences that fold to the structure while maintaining the general fold of the protein. Backbone flexibility is especially important in protein redesign because sequence mutations often result in small changes to the backbone structure. Moreover, backbone flexibility can be essential for more advanced applications of protein design, such as binding prediction and enzyme design. Some models of protein design backbone flexibility include small and continuous global backbone movements, discrete backbone samples around the target fold, backrub motions, and protein loop flexibility.Energy function
 Rational protein design techniques must be able to discriminate sequences that will be stable under the target fold from those that would prefer other low-energy competing states. Thus, protein design requires accurate energy functions that can rank and score sequences by how well they fold to the target structure. At the same time, however, these energy functions must consider the computational challenges behind protein design. One of the most challenging requirements for successful design is an energy function that is both accurate and simple for computational calculations.
The most accurate energy functions are those based on quantum mechanical simulations. However, such simulations are too slow and typically impractical for protein design. Instead, many protein design algorithms use either physics-based energy functions adapted from
Rational protein design techniques must be able to discriminate sequences that will be stable under the target fold from those that would prefer other low-energy competing states. Thus, protein design requires accurate energy functions that can rank and score sequences by how well they fold to the target structure. At the same time, however, these energy functions must consider the computational challenges behind protein design. One of the most challenging requirements for successful design is an energy function that is both accurate and simple for computational calculations.
The most accurate energy functions are those based on quantum mechanical simulations. However, such simulations are too slow and typically impractical for protein design. Instead, many protein design algorithms use either physics-based energy functions adapted from molecular mechanics
Molecular mechanics uses classical mechanics to model molecular systems. The Born–Oppenheimer approximation is assumed valid and the potential energy of all systems is calculated as a function of the nuclear coordinates using Force field (chemi ...
simulation programs, knowledge based energy-functions, or a hybrid mix of both. The trend has been toward using more physics-based potential energy functions.
Physics-based energy functions, such as AMBER
Amber is fossilized tree resin. Examples of it have been appreciated for its color and natural beauty since the Neolithic times, and worked as a gemstone since antiquity."Amber" (2004). In Maxine N. Lurie and Marc Mappen (eds.) ''Encyclopedia ...
and CHARMM
Chemistry at Harvard Macromolecular Mechanics (CHARMM) is the name of a widely used set of force fields for molecular dynamics, and the name for the molecular dynamics simulation and analysis computer software package associated with them. The CH ...
, are typically derived from quantum mechanical simulations, and experimental data from thermodynamics, crystallography, and spectroscopy. These energy functions typically simplify physical energy function and make them pairwise decomposable, meaning that the total energy of a protein conformation can be calculated by adding the pairwise energy between each atom pair, which makes them attractive for optimization algorithms. Physics-based energy functions typically model an attractive-repulsive Lennard-Jones
Sir John Edward Lennard-Jones (27 October 1894 – 1 November 1954) was a British mathematician and professor of theoretical physics at the University of Bristol, and then of theoretical chemistry, theoretical science at the University of C ...
term between atoms and a pairwise electrostatics
Electrostatics is a branch of physics that studies slow-moving or stationary electric charges.
Since classical antiquity, classical times, it has been known that some materials, such as amber, attract lightweight particles after triboelectric e ...
coulombic term between non-bonded atoms.
 Statistical potentials, in contrast to physics-based potentials, have the advantage of being fast to compute, of accounting implicitly of complex effects and being less sensitive to small changes in the protein structure. These energy functions are based on deriving energy values from frequency of appearance on a structural database.
Protein design, however, has requirements that can sometimes be limited in molecular mechanics force-fields. Molecular mechanics force-fields, which have been
used mostly in molecular dynamics simulations, are optimized for the simulation of single sequences, but protein design searches through many conformations of many sequences. Thus, molecular mechanics force-fields must be tailored for protein design. In practice, protein design energy functions often incorporate both statistical terms and physics-based terms. For example, the Rosetta energy function, one of the most-used energy functions, incorporates physics-based energy terms originating in the CHARMM energy function, and statistical energy terms, such as rotamer probability and knowledge-based electrostatics. Typically, energy functions are highly customized between laboratories, and specifically tailored for every design.
Statistical potentials, in contrast to physics-based potentials, have the advantage of being fast to compute, of accounting implicitly of complex effects and being less sensitive to small changes in the protein structure. These energy functions are based on deriving energy values from frequency of appearance on a structural database.
Protein design, however, has requirements that can sometimes be limited in molecular mechanics force-fields. Molecular mechanics force-fields, which have been
used mostly in molecular dynamics simulations, are optimized for the simulation of single sequences, but protein design searches through many conformations of many sequences. Thus, molecular mechanics force-fields must be tailored for protein design. In practice, protein design energy functions often incorporate both statistical terms and physics-based terms. For example, the Rosetta energy function, one of the most-used energy functions, incorporates physics-based energy terms originating in the CHARMM energy function, and statistical energy terms, such as rotamer probability and knowledge-based electrostatics. Typically, energy functions are highly customized between laboratories, and specifically tailored for every design.
Challenges for effective design energy functions
Water makes up most of the molecules surrounding proteins and is the main driver of protein structure. Thus, modeling the interaction between water and protein is vital in protein design. The number of water molecules that interact with a protein at any given time is huge and each one has a large number of degrees of freedom and interaction partners. Instead, protein design programs model most of such water molecules as a continuum, modeling both the hydrophobic effect and solvation polarization. Individual water molecules can sometimes have a crucial structural role in the core of proteins, and in protein–protein or protein–ligand interactions. Failing to model such waters can result in mispredictions of the optimal sequence of a protein–protein interface. As an alternative, water molecules can be added to rotamers.As an optimization problem
 The goal of protein design is to find a protein sequence that will fold to a target structure. A protein design algorithm must, thus, search all the conformations of each sequence, with respect to the target fold, and rank sequences according to the lowest-energy conformation of each one, as determined by the protein design energy function. Thus, a typical input to the protein design algorithm is the target fold, the sequence space, the structural flexibility, and the energy function, while the output is one or more sequences that are predicted to fold stably to the target structure.
The number of candidate protein sequences, however, grows exponentially with the number of protein residues; for example, there are 20100 protein sequences of length 100. Furthermore, even if amino acid side-chain conformations are limited to a few rotamers (see Structural flexibility), this results in an exponential number of conformations for each sequence. Thus, in our 100 residue protein, and assuming that each amino acid has exactly 10 rotamers, a search algorithm that searches this space will have to search over 200100 protein conformations.
The most common energy functions can be decomposed into pairwise terms between rotamers and amino acid types, which casts the problem as a combinatorial one, and powerful optimization algorithms can be used to solve it. In those cases, the total energy of each conformation belonging to each sequence can be formulated as a sum of individual and pairwise terms between residue positions. If a designer is interested only in the best sequence, the protein design algorithm only requires the lowest-energy conformation of the lowest-energy sequence. In these cases, the amino acid identity of each rotamer can be ignored and all rotamers belonging to different amino acids can be treated the same. Let ri be a rotamer at residue position i in the protein chain, and E(ri) the potential energy between the internal atoms of the rotamer. Let E(ri, rj) be the potential energy between ri and rotamer rj at residue position j. Then, we define the optimization problem as one of finding the conformation of minimum energy (ET):
The problem of minimizing ET is an
The goal of protein design is to find a protein sequence that will fold to a target structure. A protein design algorithm must, thus, search all the conformations of each sequence, with respect to the target fold, and rank sequences according to the lowest-energy conformation of each one, as determined by the protein design energy function. Thus, a typical input to the protein design algorithm is the target fold, the sequence space, the structural flexibility, and the energy function, while the output is one or more sequences that are predicted to fold stably to the target structure.
The number of candidate protein sequences, however, grows exponentially with the number of protein residues; for example, there are 20100 protein sequences of length 100. Furthermore, even if amino acid side-chain conformations are limited to a few rotamers (see Structural flexibility), this results in an exponential number of conformations for each sequence. Thus, in our 100 residue protein, and assuming that each amino acid has exactly 10 rotamers, a search algorithm that searches this space will have to search over 200100 protein conformations.
The most common energy functions can be decomposed into pairwise terms between rotamers and amino acid types, which casts the problem as a combinatorial one, and powerful optimization algorithms can be used to solve it. In those cases, the total energy of each conformation belonging to each sequence can be formulated as a sum of individual and pairwise terms between residue positions. If a designer is interested only in the best sequence, the protein design algorithm only requires the lowest-energy conformation of the lowest-energy sequence. In these cases, the amino acid identity of each rotamer can be ignored and all rotamers belonging to different amino acids can be treated the same. Let ri be a rotamer at residue position i in the protein chain, and E(ri) the potential energy between the internal atoms of the rotamer. Let E(ri, rj) be the potential energy between ri and rotamer rj at residue position j. Then, we define the optimization problem as one of finding the conformation of minimum energy (ET):
The problem of minimizing ET is an NP-hard
In computational complexity theory, a computational problem ''H'' is called NP-hard if, for every problem ''L'' which can be solved in non-deterministic polynomial-time, there is a polynomial-time reduction from ''L'' to ''H''. That is, assumi ...
problem. Even though the class of problems is NP-hard, in practice many instances of protein design can be solved exactly or optimized satisfactorily through heuristic methods.
Algorithms
Several algorithms have been developed specifically for the protein design problem. These algorithms can be divided into two broad classes: exact algorithms, such asdead-end elimination
The dead-end elimination algorithm (DEE) is a method for minimizing a function over a discrete set of independent variables. The basic idea is to identify "dead ends", i.e., combinations of variables that are not necessary to define a global mi ...
, that lack runtime guarantees but guarantee the quality of the solution; and heuristic
A heuristic or heuristic technique (''problem solving'', '' mental shortcut'', ''rule of thumb'') is any approach to problem solving that employs a pragmatic method that is not fully optimized, perfected, or rationalized, but is nevertheless ...
algorithms, such as Monte Carlo, that are faster than exact algorithms but have no guarantees on the optimality of the results. Exact algorithms guarantee that the optimization process produced the optimal according to the protein design model. Thus, if the predictions of exact algorithms fail when these are experimentally validated, then the source of error can be attributed to the energy function, the allowed flexibility, the sequence space or the target structure (e.g., if it cannot be designed for).
Some protein design algorithms are listed below. Although these algorithms address only the most basic formulation of the protein design problem, Equation (), when the optimization goal changes because designers introduce improvements and extensions to the protein design model, such as improvements to the structural flexibility allowed (e.g., protein backbone flexibility) or including sophisticated energy terms, many of the extensions on protein design that improve modeling are built atop these algorithms. For example, Rosetta Design incorporates sophisticated energy terms, and backbone flexibility using Monte Carlo as the underlying optimizing algorithm. OSPREY's algorithms build on the dead-end elimination algorithm and A* to incorporate continuous backbone and side-chain movements. Thus, these algorithms provide a good perspective on the different kinds of algorithms available for protein design.
In 2020 scientists reported the development of an AI-based process using genome databases
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
for evolution-based designing of novel proteins. They used deep learning
Deep learning is a subset of machine learning that focuses on utilizing multilayered neural networks to perform tasks such as classification, regression, and representation learning. The field takes inspiration from biological neuroscience a ...
to identify design-rules. In 2022, a study reported deep learning software that can design proteins that contain prespecified functional sites.
With mathematical guarantees
Dead-end elimination
The dead-end elimination (DEE) algorithm reduces the search space of the problem iteratively by removing rotamers that can be provably shown to be not part of the global lowest energy conformation (GMEC). On each iteration, the dead-end elimination algorithm compares all possible pairs of rotamers at each residue position, and removes each rotamer r′i that can be shown to always be of higher energy than another rotamer ri and is thus not part of the GMEC: : Other powerful extensions to the dead-end elimination algorithm include the pairs elimination criterion, and the generalized dead-end elimination criterion. This algorithm has also been extended to handle continuous rotamers with provable guarantees. Although the Dead-end elimination algorithm runs in polynomial time on each iteration, it cannot guarantee convergence. If, after a certain number of iterations, the dead-end elimination algorithm does not prune any more rotamers, then either rotamers have to be merged or another search algorithm must be used to search the remaining search space. In such cases, the dead-end elimination acts as a pre-filtering algorithm to reduce the search space, while other algorithms, such as A*, Monte Carlo, Linear Programming, or FASTER are used to search the remaining search space.Branch and bound
The protein design conformational space can be represented as atree
In botany, a tree is a perennial plant with an elongated stem, or trunk, usually supporting branches and leaves. In some usages, the definition of a tree may be narrower, e.g., including only woody plants with secondary growth, only ...
, where the protein residues are ordered in an arbitrary way, and the tree branches at each of the rotamers in a residue. Branch and bound
Branch and bound (BB, B&B, or BnB) is a method for solving optimization problems by breaking them down into smaller sub-problems and using a bounding function to eliminate sub-problems that cannot contain the optimal solution.
It is an algorithm ...
algorithms use this representation to efficiently explore the conformation tree: At each ''branching'', branch and bound algorithms ''bound'' the conformation space and explore only the promising branches.
A popular search algorithm for protein design is the A* search algorithm. A* computes a lower-bound score on each partial tree path that lower bounds (with guarantees) the energy of each of the expanded rotamers. Each partial conformation is added to a priority queue and at each iteration the partial path with the lowest lower bound is popped from the queue and expanded. The algorithm stops once a full conformation has been enumerated and guarantees that the conformation is the optimal.
The A* score f in protein design consists of two parts, f=g+h. g is the exact energy of the rotamers that have already been assigned in the partial conformation. h is a lower bound on the energy of the rotamers that have not yet been assigned. Each is designed as follows, where d is the index of the last assigned residue in the partial conformation.
:
: