|
USP4
Ubiquitin specific protease 4 (USP4) is an enzyme that cleaves ubiquitin from a number of protein substrates. Prior to the standardization of nomenclature USP4 was known as UNP, and was one of the first deubiquitinating enzymes to be identified in mammals. In the mouse and human the USP4 protein is encoded by a gene containing 22 exons. This protein is a member of cysteine peptidase family C19. As a deubiquitinating enzyme it is unusual in having the capacity to cleave ubiquitin- proline bonds. This property may reflect structural flexibility in the active site of the enzyme, and may explain its ability to cleave ubiquitin chains of various linkages. USP4 has substrates of important function in a number of cell signalling pathways, including the NF-κB, TGF-β, Wnt/β-catenin, p53, and spliceosome pathways. Other substrates include the adenosine A2A receptor and the Ro52 (TRIM21) protein. USP4 is a nucleocytoplasmic shuttling protein that bears a functional nuclear localizat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deubiquitinating Enzyme
Deubiquitinating enzymes (DUBs), also known as deubiquitinating peptidases, deubiquitinating isopeptidases, deubiquitinases, ubiquitin proteases, ubiquitin hydrolases, ubiquitin isopeptidases, are a large group of proteases that cleave ubiquitin from proteins. Ubiquitin is attached to proteins in order to regulate the degradation of proteins via the proteasome and lysosome; coordinate the cellular localisation of proteins; activate and inactivate proteins; and modulate protein-protein interactions. DUBs can reverse these effects by cleaving the peptide or isopeptide bond between ubiquitin and its substrate protein. In humans there are nearly 100 DUB genes, which can be classified into two main classes: cysteine proteases and metalloproteases. The cysteine proteases comprise ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), Machado-Josephin domain proteases (MJDs) and ovarian tumour proteases (OTU). The metalloprotease group contains only the Jab1/Mov34/ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Retinoblastoma Protein
The retinoblastoma protein (protein name abbreviated pRb; gene name abbreviated ''Rb'', ''RB'' or ''RB1'') is a proto-oncogenic tumor suppressor protein that is dysfunctional in several major cancers. One function of pRb is to prevent excessive cell growth by inhibiting cell cycle progression until a cell is ready to divide. When the cell is ready to divide, pRb is phosphorylated, inactivating it, and the cell cycle is allowed to progress. It is also a recruiter of several chromatin remodeling enzymes such as methylases and acetylases. pRb belongs to the pocket protein family, whose members have a pocket for the functional binding of other proteins. Should an oncogenic protein, such as those produced by cells infected by high-risk types of human papillomavirus, bind and inactivate pRb, this can lead to cancer. The ''RB'' gene may have been responsible for the evolution of multicellularity in several lineages of life including animals. Name and genetics In humans, the pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are Ribozyme, catalytic RNA molecules, called ribozymes. Enzymes' Chemical specificity, specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine A2A Receptor
The adenosine A2A receptor, also known as ADORA2A, is an adenosine receptor, and also denotes the human gene encoding it. Structure This protein is a member of the G protein-coupled receptor (GPCR) family which possess seven transmembrane alpha helices, as well as an extracellular N-terminus and an intracellular C-terminus. Furthermore, located in the intracellular side close to the membrane is a small alpha helix, often referred to as helix 8 (H8). The crystallographic structure of the adenosine A2A receptor reveals a ligand binding pocket distinct from that of other structurally determined GPCRs (i.e., the beta-2 adrenergic receptor and rhodopsin).; Below this primary ( orthosteric) binding pocket lies a secondary ( allosteric) binding pocket. The crystal-structure of A2A bound to the antagonist ZM241385 (PDB code: 4EIY) showed that a sodium-ion can be found in this location of the protein, thus giving it the name 'sodium-ion binding pocket'. Heteromers The actions of the A ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Export Signal
A nuclear export signal (NES) is a short target peptide containing 4 hydrophobic residues in a protein that targets it for export from the cell nucleus to the cytoplasm through the nuclear pore complex using nuclear transport. It has the opposite effect of a nuclear localization signal, which targets a protein located in the cytoplasm for import to the nucleus. The NES is recognized and bound by exportins. NESs serve several vital cellular functions. They assist in regulating the position of proteins within the cell. Through this NESs affect transcription and several other nuclear functions that are essential to proper cell function. The export of many types of RNA from the nucleus is required for proper cellular function. The NES determines what type of pathway the varying types of RNA may use to exit the nucleus and perform their function and the NESs may effect the directionality of molecules exiting the nucleus. Structure Computer analysis of known NESs found the most ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Localization Sequence
A nuclear localization signal ''or'' sequence (NLS) is an amino acid sequence that 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines or arginines exposed on the protein surface. Different nuclear localized proteins may share the same NLS. An NLS has the opposite function of a nuclear export signal (NES), which targets proteins out of the nucleus. Types Classical These types of NLSs can be further classified as either monopartite or bipartite. The major structural differences between the two are that the two basic amino acid clusters in bipartite NLSs are separated by a relatively short spacer sequence (hence bipartite - 2 parts), while monopartite NLSs are not. The first NLS to be discovered was the sequence PKKKRKV in the SV40 Large T-antigen (a monopartite NLS). The NLS of nucleoplasmin, KR AATKKAGQAKKK, is the prototype of the ubiquitous bipartite signal: two clu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
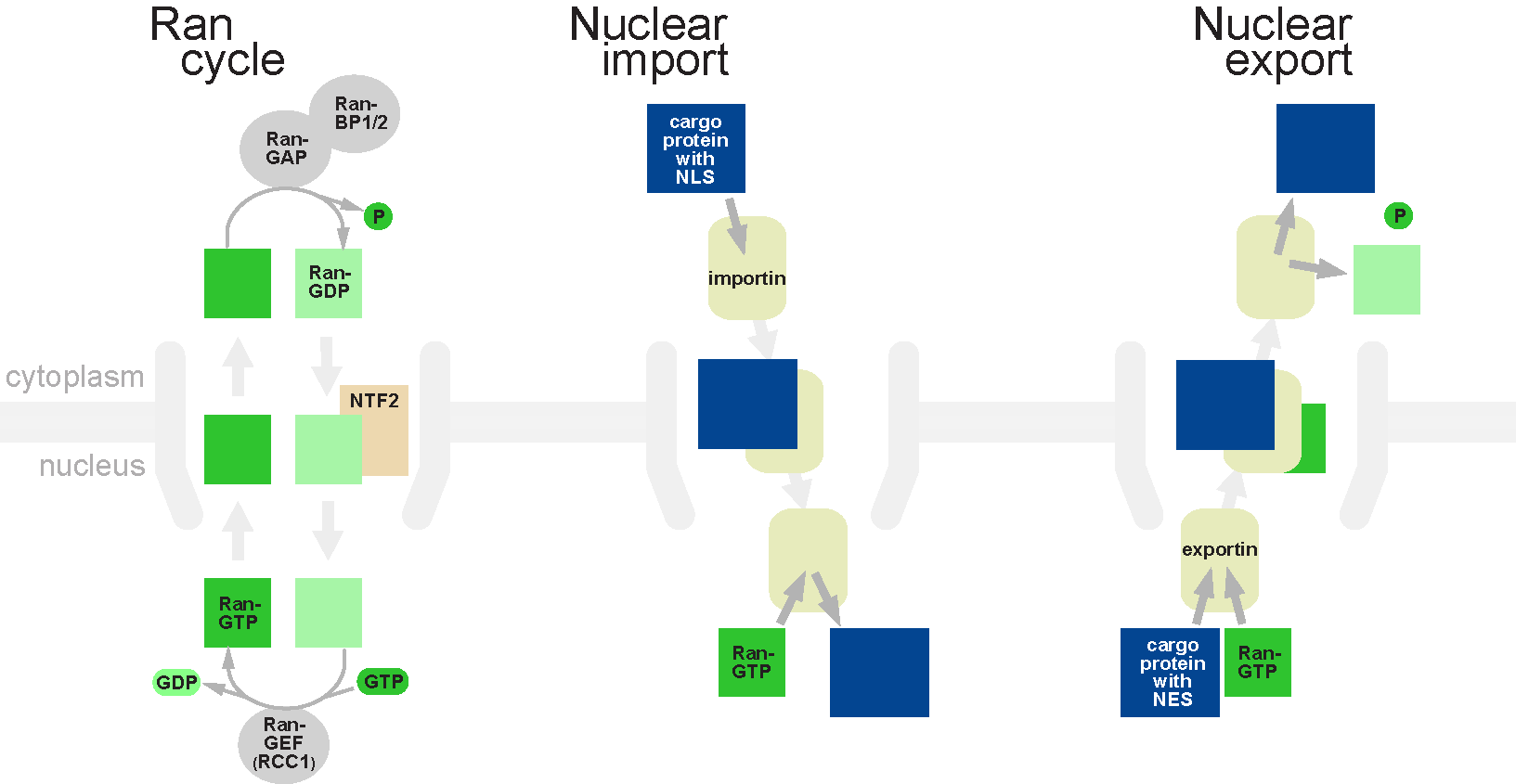
Nucleocytoplasmic Shuttling Protein
Nuclear transport refers to the mechanisms by which molecules move across the nuclear membrane of a cell. The entry and exit of large molecules from the cell nucleus is tightly controlled by the nuclear pore complexes (NPCs). Although small molecules can enter the nucleus without regulation, macromolecules such as RNA and proteins require association with transport factors known as nuclear transport receptors, like karyopherins called importins to enter the nucleus and exportins to exit. Nuclear import Protein that must be imported to the nucleus from the cytoplasm carry nuclear localization signals (NLS) that are bound by importins. An NLS is a sequence of amino acids that acts as a tag. They are most commonly hydrophilic proteins containing lysine and arginine residues, although diverse NLS sequences have been documented. Proteins, transfer RNA, and assembled ribosomal subunits are exported from the nucleus due to association with exportins, which bind signaling sequences called ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TRIM21
Tripartite motif-containing protein 21, also known as E3 ubiquitin-protein ligase TRIM21, is a protein that in humans is encoded by the ''TRIM21'' gene. Alternatively spliced transcript variants for this gene have been described but the full-length nature of only one has been determined. It is expressed in most human tissues. Structure TRIM21 is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING finger domain, a B-box type 1 and a B-box type 2 zinc finger, and a coiled coil region. Function TRIM21 is an intracellular antibody effector in the intracellular antibody-mediated proteolysis pathway. It recognizes Fc domain and binds to immunoglobulin G, immunoglobulin A and immunoglobulin M on antibody marked non-enveloped virions which have infected the cell. Either by autoubiquitination or by ubiquitination of a cofactor, it is then responsible for directing the virions to the proteasome. TRIM21 itself is not degraded in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Spliceosome
A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs ( snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to specific proteins to form a small nuclear ribonucleoprotein complex (snRNP, pronounced “snurps”), which in turn combines with other snRNPs to form a large ribonucleoprotein complex called a spliceosome. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material (equivalent to the introns) from the initial film and sends the cleaned-up version to the director for the final cut. However, sometimes the RNA within the intron acts as a ribozyme, splicing itself without the use of a spliceosome or protein enzymes. History In 1977, work by the Sharp and Roberts labs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or, alternatively, ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Wnt Signaling Pathway
The Wnt signaling pathways are a group of signal transduction pathways which begin with proteins that pass signals into a cell through cell surface receptors. The name Wnt is a portmanteau created from the names Wingless and Int-1. Wnt signaling pathways use either nearby cell-cell communication (paracrine) or same-cell communication (autocrine). They are highly evolutionarily conserved in animals, which means they are similar across animal species from fruit flies to humans. Three Wnt signaling pathways have been characterized: the canonical Wnt pathway, the noncanonical planar cell polarity pathway, and the noncanonical Wnt/calcium pathway. All three pathways are activated by the binding of a Wnt-protein ligand to a Frizzled family receptor, which passes the biological signal to the Dishevelled protein inside the cell. The canonical Wnt pathway leads to regulation of gene transcription, and is thought to be negatively regulated in part by the SPATS1 gene. The noncanonical ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |