|
TWAS V1
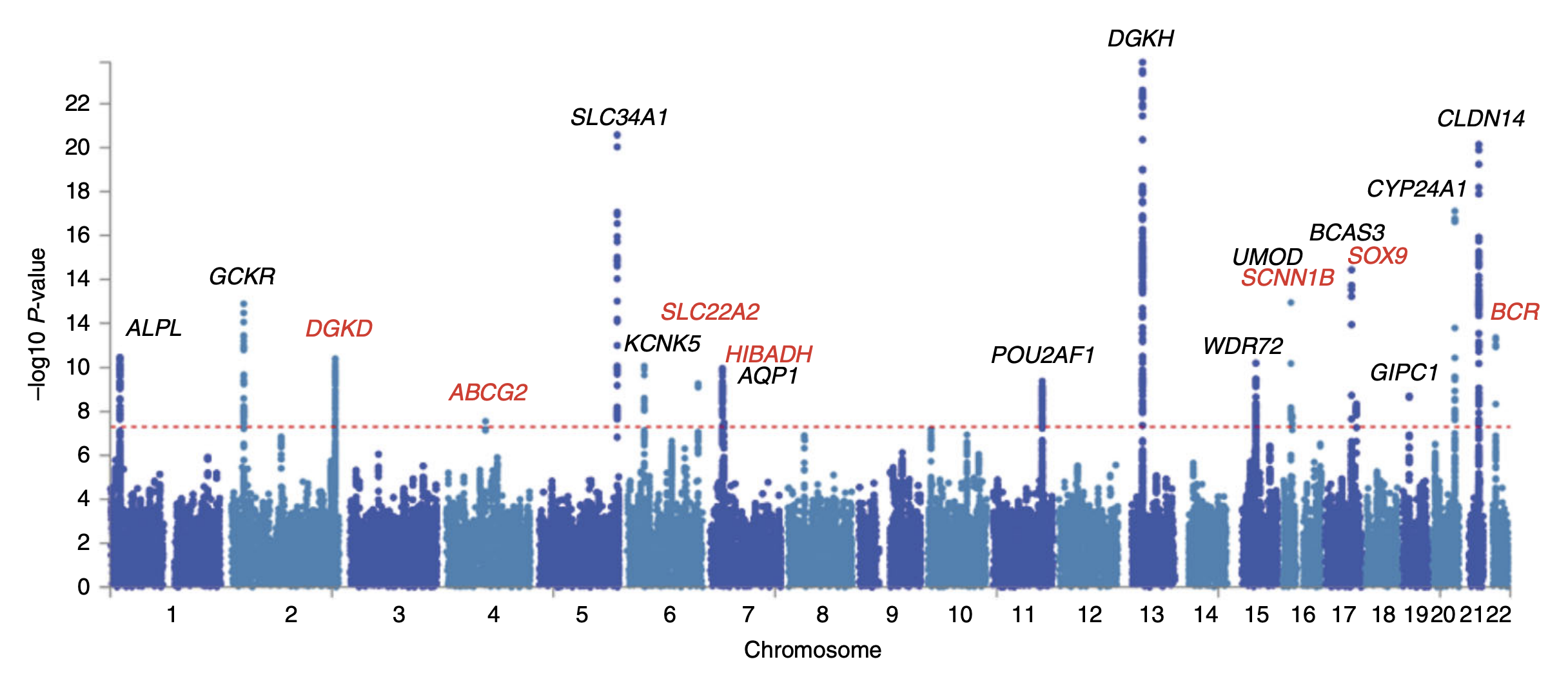
Transcriptome-wide association study (TWAS) is a statistical genetics methodology to improve detection power and provide functional annotation for genetic associations with phenotypes by integrating single-nucleotide polymorphism to trait (SNP-trait) associations from genome-wide association studies with SNP-based prediction models of gene expression. The approach was presented by Eric R. Gamazon et al. and Alexander Gusev et al. in the journal ''Nature Genetics''. This methodology has been widely adopted, having received 2057 citations (as of December 24, 2021) according to Google Scholar. See also * Genome-wide association study * Functional genomics * Genotype-Tissue Expression (GTEx) project * Epigenome-wide association study An epigenome-wide association study (EWAS) is an examination of a genome-wide set of quantifiable epigenetic marks, such as DNA methylation, in different individuals to derive associations between epigenetic variation and a particular identifiab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Statistical Genetics
Statistical genetics is a scientific field concerned with the development and application of statistical methods for drawing inferences from genetic data. The term is most commonly used in the context of human genetics. Research in statistical genetics generally involves developing theory or methodology to support research in one of three related areas: *population genetics - Study of evolutionary processes affecting genetic variation between organisms *genetic epidemiology - Studying effects of genes on diseases *quantitative genetics - Studying the effects of genes on 'normal' phenotypes Statistical geneticists tend to collaborate closely with geneticists, molecular biologists, clinicians and bioinformaticians. Statistical genetics is a type of computational biology Computational biology refers to the use of data analysis, mathematical modeling and computational simulations to understand biological systems and relationships. An intersection of computer science, biology, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphism
In genetics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, at a specific base position in the human genome, the G nucleotide may appear in most individuals, but in a minority of individuals, the position is occupied by an A. This means that there is a SNP at this specific position, and the two possible nucleotide variations – G or A – are said to be the alleles for this specific position. SNPs pinpoint differences in our susceptibility to a wide range of diseases, for example age-related macular degeneration (a common SNP in the CFH gene is associated with increased risk of the disease) or nonalcoholic fatty liver disease (a SNP in the PNPLA3 gene is associated with inc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome-wide Association Study
In genomics, a genome-wide association study (GWA study, or GWAS), also known as whole genome association study (WGA study, or WGAS), is an observational study of a genome-wide set of Single-nucleotide polymorphism, genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms. When applied to human data, GWA studies compare the DNA of participants having varying phenotypes for a particular trait or disease. These participants may be people with a disease (cases) and similar people without the disease (controls), or they may be people with different phenotypes for a particular trait, for example blood pressure. This approach is known as phenotype-first, in which the participants are classified first by their clinical manifestation(s), as oppose ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eric R
The given name Eric, Erich, Erikk, Erik, Erick, or Eirik is derived from the Old Norse name ''Eiríkr'' (or ''Eríkr'' in Old East Norse due to monophthongization). The first element, ''ei-'' may be derived from the older Proto-Norse ''* aina(z)'', meaning "one, alone, unique", ''as in the form'' ''Æ∆inrikr'' explicitly, but it could also be from ''* aiwa(z)'' "everlasting, eternity", as in the Gothic form ''Euric''. The second element ''- ríkr'' stems either from Proto-Germanic ''* ríks'' "king, ruler" (cf. Gothic ''reiks'') or the therefrom derived ''* ríkijaz'' "kingly, powerful, rich, prince"; from the common Proto-Indo-European root * h₃rḗǵs. The name is thus usually taken to mean "sole ruler, autocrat" or "eternal ruler, ever powerful". ''Eric'' used in the sense of a proper noun meaning "one ruler" may be the origin of ''Eriksgata'', and if so it would have meant "one ruler's journey". The tour was the medieval Swedish king's journey, when newly elected, to s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alexander Gusev (scientist)
Alexander (Sasha) Gusev is a computational biologist and an Assistant Professor of Medicine at Harvard Medical School. Research and career Alexander Gusev has developed computational methods that use genetic data to decipher disease mechanisms. For example, he has identified 34 new genes associated with increased risk of earliest-stage ovarian cancer. He has developed computational methods that integrate molecular data to facilitate functional interpretation of findings from genome-wide association studies. He has contributed to the development of the transcriptome-wide association study approach to mapping disease-associated genes. In addition, he studies the interactions between germline In biology and genetics, the germline is the population of a multicellular organism's cells that pass on their genetic material to the progeny (offspring). In other words, they are the cells that form the egg, sperm and the fertilised egg. They ... (host) and somatic events (tumor) - which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nature Genetics
''Nature Genetics'' is a peer-reviewed scientific journal published by Nature Portfolio. It was established in 1992. It covers research in genetics. The chief editor is Tiago Faial. The journal encompasses genetic and functional genomic studies on human traits and on other model organisms, including mouse, fly, nematode and yeast. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation. According to the ''Journal Citation Reports'', the journal has a 2021 impact factor The impact factor (IF) or journal impact factor (JIF) of an academic journal is a scientometric index calculated by Clarivate that reflects the yearly mean number of citations of articles published in the last two years in a given journal, as ... of 41.379, ranking it 2nd out of 175 journals in the category "Genetics & Heredity". References External links Official websit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Functional Genomics
Functional genomics is a field of molecular biology that attempts to describe gene (and protein) functions and interactions. Functional genomics make use of the vast data generated by genomic and transcriptomic projects (such as genome sequencing projects and RNA sequencing). Functional genomics focuses on the dynamic aspects such as gene transcription, translation, regulation of gene expression and protein–protein interactions, as opposed to the static aspects of the genomic information such as DNA sequence or structures. A key characteristic of functional genomics studies is their genome-wide approach to these questions, generally involving high-throughput methods rather than a more traditional "gene-by-gene" approach. Definition and goals of functional genomics In order to understand functional genomics it is important to first define function. In their paper Graur et al. define function in two possible ways. These are "selected effect" and "causal role". The "selected ef ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenome-wide Association Study
An epigenome-wide association study (EWAS) is an examination of a genome-wide set of quantifiable epigenetic marks, such as DNA methylation, in different individuals to derive associations between epigenetic variation and a particular identifiable phenotype/trait. When patterns change such as DNA methylation at specific loci, discriminating the phenotypically affected cases from control individuals, this is considered an indication that epigenetic perturbation has taken place that is associated, causally or consequentially, with the phenotype. Background The epigenome is governed by both genetic and environmental factors, causing it to be highly dynamic and complex. Epigenetic information exists in the cell as DNA and histone marks, as well as non-coding RNAs. DNA methylation (DNAm) patterns change over time, and vary between developmental stage and tissue type. The main type of DNAm is at cytosines within CpG dinucleotides which is known to be involved in gene expression reg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |