|
TCHH
Trichohyalin is a protein that in mammals is encoded by the ''TCHH'' gene. Discovery In 1903 the name ''trichohyalin'' was assigned to the granules of the inner root sheath (IRS) of hair follicles discovered by Hans Vörner. In 1986 the name was reassigned to a protein isolated from sheep wool follicles. Gene location The human TCHH is located on the long (q) arm of chromosome 1 at region 2 band 1 sub-band 3 (1q21.3), from base pair 152,105,403 to base pair 152,116,368map. This region in chromosome 1q21 is known as the epidermal differentiation complex, since it harbors over fifty other genes involved in keratinocyte differentiation. Gene coding sequence contains 5829 nucleotides. Gene orthologs were identified in most mammals including mice, chickens, rats, pigs, sheep, horses and other species. Protein localisation Trichohyalin is highly expressed in the inner root sheath cells of the hair follicle and medulla. It was also detected in the granular layer and stratum corne ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epidermal Differentiation Complex
The epidermal differentiation complex (EDC) is a gene complex comprising over fifty genes encoding proteins involved in the terminal differentiation and cornification of keratinocytes, the primary cell type of the epidermis. In humans, the complex is located on a 1.9 Mbp stretch within chromosome 1q21. The proteins encoded by EDC genes are closely related in terms of function, and evolutionarily they belong to three distinct gene families: the cornified envelope precursor family, the S100 protein family and the S100 fused type protein (SFTP) family. It has been hypothesized that the clustering of EDC genes occurred due to duplication events which were evolutionarily favored during the adaptation to terrestrial environments. EDC proteins have been involved in a variety of skin disorders including ichthyosis vulgaris, atopic dermatitis and psoriasis. History The epidermal differentiation complex was first described in 1993, and further characterized in 1996, when Dietmar Mischke an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Citrulline
The organic compound citrulline is an α-amino acid. Its name is derived from ''citrullus'', the Latin word for watermelon. Although named and described by gastroenterologists since the late 19th century, it was first isolated from watermelon in 1914 by Japanese researchers Yotaro Koga and Ryo OdakeEarly references spell Ryo Odake's name as ''Ryo Othake''. and further codified by Mitsunori Wada of Tokyo Imperial University in 1930. It has the formula H2NC(O)NH(CH2)3CH(NH2)CO2H. It is a key intermediate in the urea cycle, the pathway by which mammals excrete ammonia by converting it into urea. Citrulline is also produced as a byproduct of the enzymatic production of nitric oxide from the amino acid arginine, catalyzed by nitric oxide synthase. Biosynthesis Citrulline can be derived from: * from arginine via nitric oxide synthase, as a byproduct of the production of nitric oxide for signaling purposes * from ornithine through the breakdown of proline or glutamine/glutamate * from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EF Hand
The EF hand is a helix–loop–helix structural domain or ''motif'' found in a large family of calcium-binding proteins. The EF-hand motif contains a helix–loop–helix topology, much like the spread thumb and forefinger of the human hand, in which the Ca2+ ions are coordinated by ligands within the loop. The motif takes its name from traditional nomenclature used in describing the protein parvalbumin, which contains three such motifs and is probably involved in muscle relaxation via its calcium-binding activity. The EF-hand consists of two alpha helices linked by a short loop region (usually about 12 amino acids) that usually binds calcium ions. EF-hands also appear in each structural domain of the signaling protein calmodulin and in the muscle protein troponin-C. Calcium ion binding site The calcium ion is coordinated in a pentagonal bipyramidal configuration. The six residues involved in the binding are in positions 1, 3, 5, 7, 9 and 12; these residues are denoted by X ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
S100 Protein
The S100 proteins are a family of low molecular-weight proteins found in vertebrates characterized by two calcium-binding sites that have helix-loop-helix ("EF-hand-type") conformation. At least 21 different S100 proteins are known. They are encoded by a family of genes whose symbols use the ''S100'' prefix, for example, ''S100A1'', ''S100A2'', ''S100A3''. They are also considered as damage-associated molecular pattern molecules (DAMPs), and knockdown of aryl hydrocarbon receptor downregulates the expression of S100 proteins in THP-1 cells. Structure Most S100 proteins consist of two identical polypeptides (homodimeric), which are held together by noncovalent bonds. They are structurally similar to calmodulin. They differ from calmodulin, though, on the other features. For instance, their expression pattern is cell-specific, i.e. they are expressed in particular cell types. Their expression depends on environmental factors. In contrast, calmodulin is a ubiquitous and universa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Keratin
Keratin () is one of a family of structural fibrous proteins also known as ''scleroproteins''. Alpha-keratin (α-keratin) is a type of keratin found in vertebrates. It is the key structural material making up scales, hair, nails, feathers, horns, claws, hooves, and the outer layer of skin among vertebrates. Keratin also protects epithelial cells from damage or stress. Keratin is extremely insoluble in water and organic solvents. Keratin monomers assemble into bundles to form intermediate filaments, which are tough and form strong unmineralized epidermal appendages found in reptiles, birds, amphibians, and mammals. Excessive keratinization participate in fortification of certain tissues such as in horns of cattle and rhinos, and armadillos' osteoderm. The only other biological matter known to approximate the toughness of keratinized tissue is chitin. Keratin comes in two types, the primitive, softer forms found in all vertebrates and harder, derived forms found only amon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
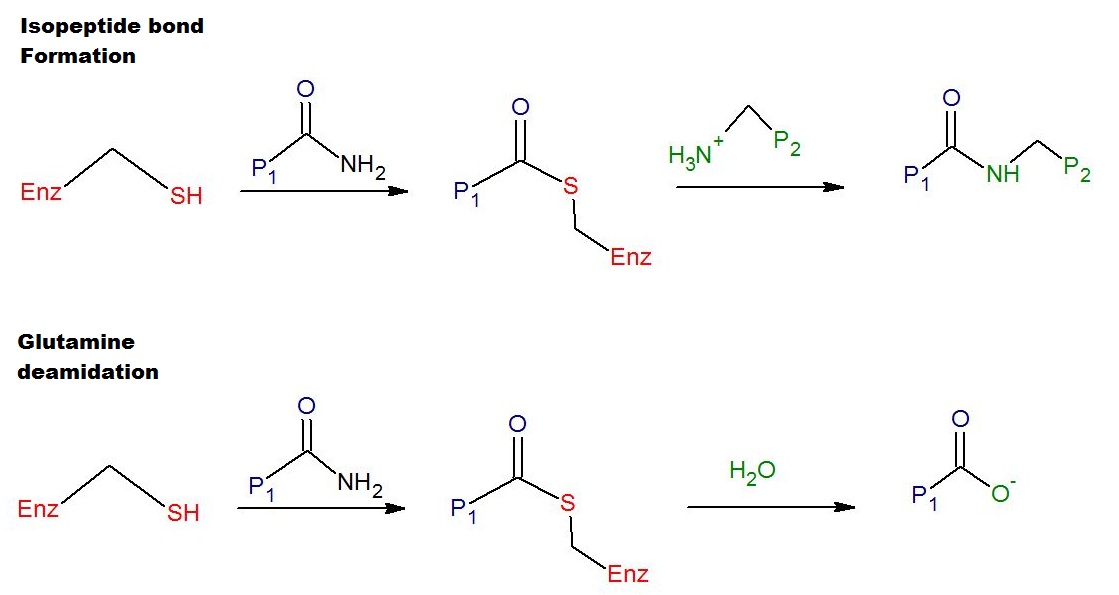
Transglutaminase
Transglutaminases are enzymes that in nature primarily catalyze the formation of an isopeptide bond between γ-carboxamide groups ( -(C=O)NH2 ) of glutamine residue side chains and the ε-amino groups ( -NH2 ) of lysine residue side chains with subsequent release of ammonia ( NH3 ). Lysine and glutamine residues must be bound to a peptide or a protein so that this cross-linking (between separate molecules) or intramolecular (within the same molecule) reaction can happen. Bonds formed by transglutaminase exhibit high resistance to proteolytic degradation (proteolysis). The reaction is :Glutamine, Gln-(C=O)NH2 + Lysine, NH2-Lys → Gln-(C=O)NH-Lys + NH3 Transglutaminases can also join a primary amine ( RNH2 ) to the side chain carboxyamide group of a protein/peptide bound glutamine residue thus forming an isopeptide bond :Gln-(C=O)NH2 + RNH2 → Gln-(C=O)NHR + NH3 These enzymes can also Deamidation, deamidate glutamine residues to glutamic acid residues ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deimination
Citrullination or deimination is the conversion of the amino acid arginine in a protein into the amino acid citrulline. Citrulline is not one of the 20 standard amino acids encoded by DNA in the genetic code. Instead, it is the result of a post-translational modification. Citrullination is distinct from the formation of the free amino acid citrulline as part of the urea cycle or as a byproduct of enzymes of the nitric oxide synthase family. Enzymes called arginine deiminases (ADIs) catalyze the deimination of free arginine, while protein arginine deiminases or peptidylarginine deiminases (PADs) replace the primary ketimine group (>C=NH) by a ketone group (>C=O). Arginine is positively charged at a neutral pH, whereas citrulline has no net charge. This increases the hydrophobicity of the protein, which can lead to changes in protein folding, affecting the structure and function. The immune system can attack citrullinated proteins, leading to autoimmune diseases such as rheumatoid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptidylarginine Deiminases
In enzymology, a protein-arginine deiminase () is an enzyme that catalyzes a form of post translational modification called arginine de-imination or citrullination: :protein L-arginine + H2O \rightleftharpoons protein L-citrulline + NH3 Thus, the two substrates of this enzyme are protein L-arginine (arginine residue inside a protein) and H2O, whereas its two products are protein L-citrulline and NH3: : This enzyme belongs to the family of hydrolases, those acting on carbon-nitrogen bonds other than peptide bonds, specifically in linear amidines. The systematic name of this enzyme class is protein-L-arginine iminohydrolase. This enzyme is also called peptidylarginine deiminase. Structural studies As of late 2007, seven structures have been solved for this class of enzymes, with PDB accession codes , , , , , , and . Mammalian proteins Mammals have 5 protein-arginine deiminases, with symbols *PADI1, PADI2, PADI3, PADI4, PADI6 except for rodent Rodents (from Lat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptide Bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein chain. It can also be called a eupeptide bond to distinguish it from an isopeptide bond, which is another type of amide bond between two amino acids. Synthesis When two amino acids form a ''dipeptide'' through a ''peptide bond'', it is a type of condensation reaction. In this kind of condensation, two amino acids approach each other, with the non-side chain (C1) carboxylic acid moiety of one coming near the non-side chain (N2) amino moiety of the other. One loses a hydrogen and oxygen from its carboxyl group (COOH) and the other loses a hydrogen from its amino group (NH2). This reaction produces a molecule of water (H2O) and two amino acids joined by a peptide bond (−CO−NH−). The two joined amino acids are called a dipeptide. The am ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer aft ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |