|
SOS Box
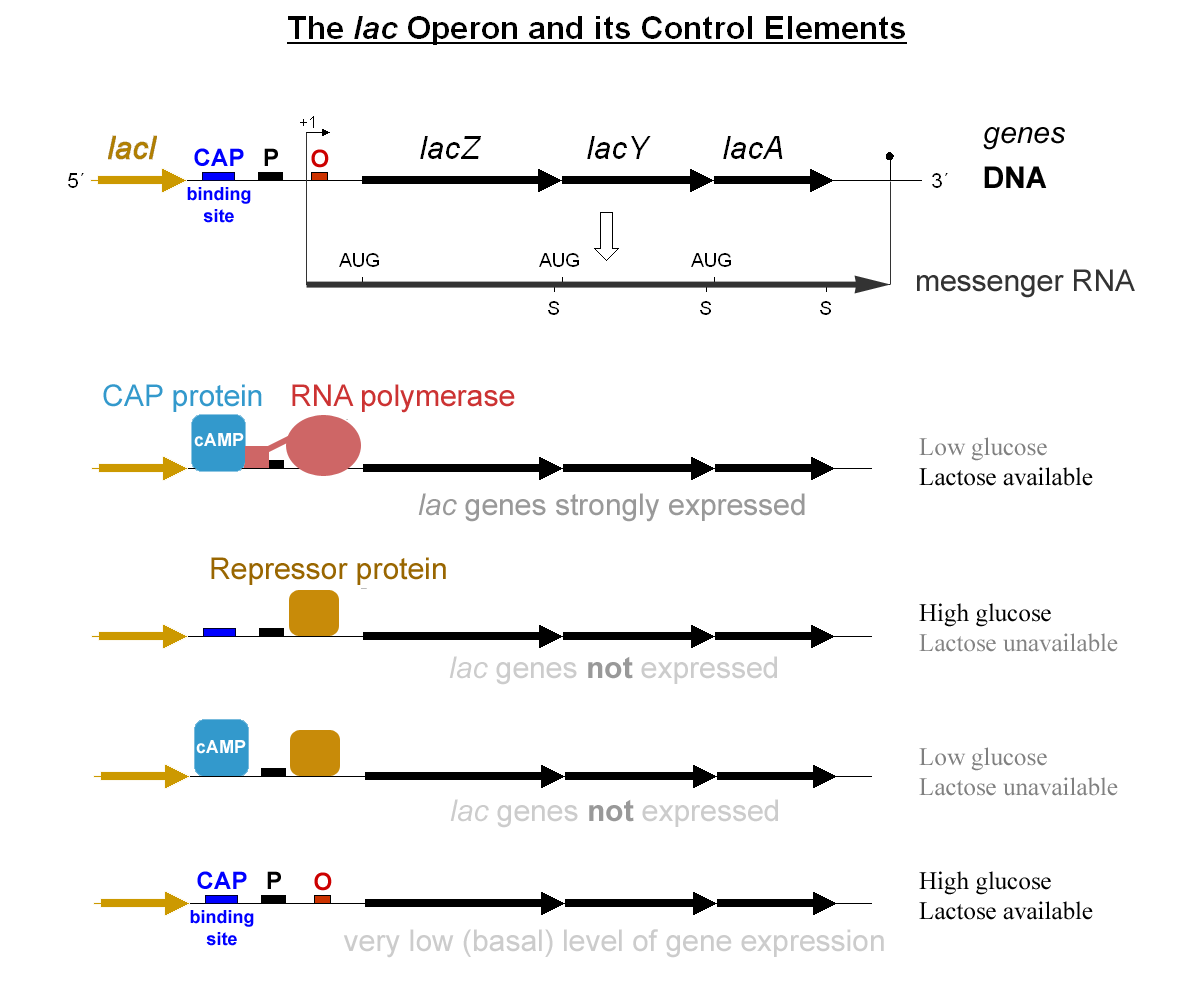
SOS box is the region in the promoter of various genes to which the LexA repressor binds to repress the transcription of SOS-induced proteins. This occurs in the absence of DNA damage. In the presence of DNA damage the binding of LexA is inactivated by the RecA activator. SOS boxes differ in DNA sequences and binding affinity towards LexA from organism to organism. Furthermore, SOS boxes may be present in a dual fashion, which indicates that more than one SOS box can be within the same promoter. Examples See Nucleic acid nomenclature for an explanation of non-GATC nucleotide letters. See also * SOS response * SOS gene * LexA * RecA RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA. A RecA structural and functional homolog has been found in every species in which one has been seriously sought and serves as an archetype for this class of homolog ... References * * DNA repair {{genetics-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promotor (biology)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Betaproteobacteria
Betaproteobacteria are a class of Gram-negative bacteria, and one of the eight classes of the phylum Pseudomonadota (synonym Proteobacteria). The ''Betaproteobacteria'' are a class comprising over 75 genera and 400 species of bacteria. Together, the ''Betaproteobacteria'' represent a broad variety of metabolic strategies and occupy diverse environments from obligate pathogens living within host organisms to oligotrophic groundwater ecosystems. Whilst most members of the ''Betaproteobacteria'' are heterotrophic, deriving both their carbon and electrons from organocarbon sources, some are photoheterotrophic, deriving energy from light and carbon from organocarbon sources. Other genera are autotrophic, deriving their carbon from bicarbonate or carbon dioxide and their electrons from reduced inorganic ions such as nitrite, ammonium, thiosulfate or sulfide — many of these chemolithoautotrophic. ''Betaproteobacteria'' are economically important, with roles in maintaining soil p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SOS Response
The SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis is induced. The system involves the RecA protein ( Rad51 in eukaryotes). The RecA protein, stimulated by single-stranded DNA, is involved in the inactivation of the repressor (LexA) of SOS response genes thereby inducing the response. It is an error-prone repair system that contributes significantly to DNA changes observed in a wide range of species. Discovery The SOS response was discovered and named by Miroslav Radman in 1975. Mechanism During normal growth, the SOS genes are negatively regulated by LexA repressor protein dimers. Under normal conditions, LexA binds to a 20-bp consensus sequence (the SOS box) in the operator region for those genes. Some of these SOS genes are expressed at certain levels even in the repressed state, according to the affinity of LexA for their SOS box. Activation of the SOS genes occurs after DNA damage by the accumulation of s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Nomenclature
The nucleic acid notation currently in use was first formalized by the International Union of Pure and Applied Chemistry (IUPAC) in 1970. This universally accepted notation uses the Roman characters G, C, A, and T, to represent the four nucleotides commonly found in deoxyribonucleic acids (DNA). Given the rapidly expanding role for genetic sequencing, synthesis, and analysis in biology, some researchers have developed alternate notations to further support the analysis and manipulation of genetic data. These notations generally exploit size, shape, and symmetry to accomplish these objectives. IUPAC notation Degenerate base symbols in biochemistry are an IUPAC representation for a position on a DNA sequence that can have multiple possible alternatives. These should not be confused with non-canonical bases because each particular sequence will have in fact one of the regular bases. These are used to encode the consensus sequence of a population of aligned sequences and are used f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cyanobacteria
Cyanobacteria (), also known as Cyanophyta, are a phylum of gram-negative bacteria that obtain energy via photosynthesis. The name ''cyanobacteria'' refers to their color (), which similarly forms the basis of cyanobacteria's common name, blue-green algae, although they are not usually scientifically classified as algae. They appear to have originated in a freshwater or terrestrial environment. Sericytochromatia, the proposed name of the paraphyletic and most basal group, is the ancestor of both the non-photosynthetic group Melainabacteria and the photosynthetic cyanobacteria, also called Oxyphotobacteria. Cyanobacteria use photosynthetic pigments, such as carotenoids, phycobilins, and various forms of chlorophyll, which absorb energy from light. Unlike heterotrophic prokaryotes, cyanobacteria have internal membranes. These are flattened sacs called thylakoids where photosynthesis is performed. Phototrophic eukaryotes such as green plants perform photosynthesis in plast ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gram-positive Bacteria
In bacteriology, gram-positive bacteria are bacteria that give a positive result in the Gram stain test, which is traditionally used to quickly classify bacteria into two broad categories according to their type of cell wall. Gram-positive bacteria take up the crystal violet stain used in the test, and then appear to be purple-coloured when seen through an optical microscope. This is because the thick peptidoglycan layer in the bacterial cell wall retains the stain after it is washed away from the rest of the sample, in the decolorization stage of the test. Conversely, gram-negative bacteria cannot retain the violet stain after the decolorization step; alcohol used in this stage degrades the outer membrane of gram-negative cells, making the cell wall more porous and incapable of retaining the crystal violet stain. Their peptidoglycan layer is much thinner and sandwiched between an inner cell membrane and a bacterial outer membrane, causing them to take up the counterstain (sa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deltaproteobacteria
The Myxococcota are a phylum of bacteria known as the fruiting gliding bacteria. All species of this group are Gram-negative. They are predominantly aerobic genera that release myxospores in unfavorable environments. Phylogeny The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LSPN) and the National Center for Biotechnology Information (NCBI). See also * List of bacterial orders * List of bacteria genera * Bacterial taxonomy Bacterial taxonomy is the taxonomy, i.e. the rank-based classification, of bacteria. In the scientific classification established by Carl Linnaeus, each species has to be assigned to a genus ( binary nomenclature), which in turn is a lower level ... References External links * {{Taxonbar, from=Q307535 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gammaproteobacteria
Gammaproteobacteria is a class of bacteria in the phylum Pseudomonadota (synonym Proteobacteria). It contains about 250 genera, which makes it the most genera-rich taxon of the Prokaryotes. Several medically, ecologically, and scientifically important groups of bacteria belong to this class. It is composed by all Gram-negative microbes and is the most phylogenetically and physiologically diverse class of Proteobacteria. These microorganisms can live in several terrestrial and marine environments, in which they play various important roles, including ''extreme environments'' such as hydrothermal vents. They generally have different shapes - rods, curved rods, cocci, spirilla, and filaments and include free living bacteria, biofilm formers, commensals and symbionts, some also have the distinctive trait of being bioluminescent. Metabolisms found in the different genera are very different; there are both aerobic and anaerobic (obligate or facultative) species, chemolithoautotrophic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alphaproteobacteria
Alphaproteobacteria is a class of bacteria in the phylum Pseudomonadota (formerly Proteobacteria). The Magnetococcales and Mariprofundales are considered basal or sister to the Alphaproteobacteria. The Alphaproteobacteria are highly diverse and possess few commonalities, but nevertheless share a common ancestor. Like all ''Proteobacteria'', its members are gram-negative and some of its intracellular parasitic members lack peptidoglycan and are consequently gram variable. Characteristics The Alphaproteobacteria are a diverse taxon and comprises several phototrophic genera, several genera metabolising C1-compounds (''e.g.'', ''Methylobacterium'' spp.), symbionts of plants (''e.g.'', ''Rhizobium'' spp.), endosymbionts of arthropods (''Wolbachia'') and intracellular pathogens (''e.g. Rickettsia''). Moreover, the class is sister to the protomitochondrion, the bacterium that was engulfed by the eukaryotic ancestor and gave rise to the mitochondria, which are organelles in eukaryotic ce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Activator (genetics)
A transcriptional activator is a protein (transcription factor) that increases transcription of a gene or set of genes. Activators are considered to have ''positive'' control over gene expression, as they function to promote gene transcription and, in some cases, are required for the transcription of genes to occur. Most activators are DNA-binding proteins that bind to enhancers or promoter-proximal elements. The DNA site bound by the activator is referred to as an "activator-binding site". The part of the activator that makes protein–protein interactions with the general transcription machinery is referred to as an "activating region" or "activation domain". Most activators function by binding sequence-specifically to a regulatory DNA site located near a promoter and making protein–protein interactions with the general transcription machinery (RNA polymerase and general transcription factors), thereby facilitating the binding of the general transcription machinery to the prom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RecA
RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA. A RecA structural and functional homolog has been found in every species in which one has been seriously sought and serves as an archetype for this class of homologous DNA repair proteins. The homologous protein is called RAD51 in eukaryotes and RadA in archaea. RecA has multiple activities, all related to DNA repair. In the bacterial SOS response, it has a co-protease function in the autocatalytic cleavage of the LexA repressor and the λ repressor. RecA's association with DNA repair is based on its central role in homologous recombination. The RecA protein binds strongly and in long clusters to ssDNA to form a nucleoprotein filament. The protein has more than one DNA binding site, and thus can hold a single strand and double strand together. This feature makes it possible to catalyze a DNA synapsis reaction between a DNA double helix and a complementary region of single-stranded DNA. The RecA-s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |