|
SNAP-tag
SNAP-tag® is a self-labeling protein tag commercially available in various expression vectors. SNAP-tag is a 182 residues polypeptide (19.4 kDa) that can be fused to any protein of interest and further specifically and covalently tagged with a suitable ligand, such as a fluorescent dye. Since its introduction, SNAP-tag has found numerous applications in biochemistry and for the investigation of the function and localisation of proteins and enzymes in living cells. Compared to the current standard labelling methods used in fluorescence microscopy, the use of SNAP-tag presents significant advantages. SNAP-tag® is a registered trademark of New England Biolabs, Inc. Applications Cell biology utilizes tools that allow manipulation and visualization of proteins in living cells. An important example is the use of fluorescent proteins, such as the green fluorescent protein (GFP) or yellow fluorescent protein (YFP). Molecular biology methods allow these fluorescent proteins to be intr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SNAP-tag
SNAP-tag® is a self-labeling protein tag commercially available in various expression vectors. SNAP-tag is a 182 residues polypeptide (19.4 kDa) that can be fused to any protein of interest and further specifically and covalently tagged with a suitable ligand, such as a fluorescent dye. Since its introduction, SNAP-tag has found numerous applications in biochemistry and for the investigation of the function and localisation of proteins and enzymes in living cells. Compared to the current standard labelling methods used in fluorescence microscopy, the use of SNAP-tag presents significant advantages. SNAP-tag® is a registered trademark of New England Biolabs, Inc. Applications Cell biology utilizes tools that allow manipulation and visualization of proteins in living cells. An important example is the use of fluorescent proteins, such as the green fluorescent protein (GFP) or yellow fluorescent protein (YFP). Molecular biology methods allow these fluorescent proteins to be intr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Tag
Protein tags are peptide sequences genetically grafted onto a recombinant protein. Tags are attached to proteins for various purposes. They can be added to either end of the target protein, so they are either C-terminus or N-terminus specific or are both C-terminus and N-terminus specific. Some tags are also inserted at sites within the protein of interest; they are known as internal tags. Affinity tags are appended to proteins so that they can be purified from their crude biological source using an affinity technique. Affinity tags include chitin binding protein (CBP), maltose binding protein (MBP), Strep-tag and glutathione-S-transferase (GST). The poly(His) tag is a widely used protein tag, which binds to matrices bearing immobilized metal ions. Solubilization tags are used, especially for recombinant proteins expressed in species such as '' E. coli'', to assist in the proper folding in proteins and keep them from aggregating in inclusion bodies. These tags include thiore ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
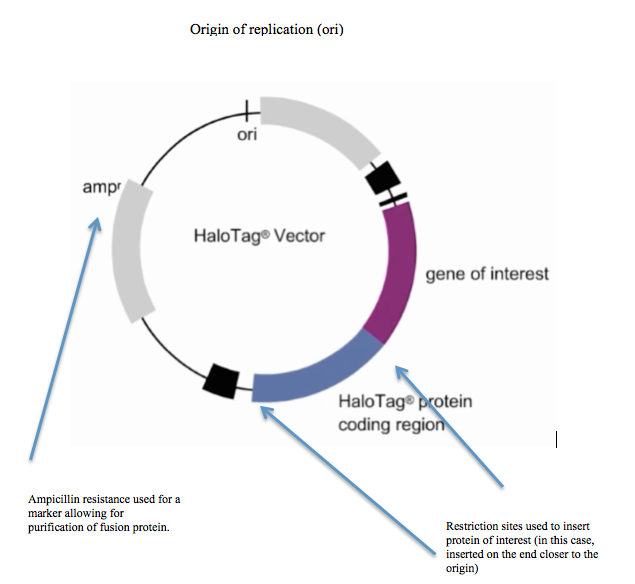
HaloTag
HaloTag is a self-labeling protein tag. It is a 297 residue protein (33 kDa) derived from a bacterial enzyme, designed to covalently bind to a synthetic ligand. The bacterial enzyme can be fused to various proteins of interest. The synthetic ligand is chosen from a number of available ligands in accordance with the type of experiments to be performed. This bacterial enzyme is a haloalkane dehalogenase, which acts as a hydrolase and is designed to facilitate visualization of the subcellular localization of a protein of interest, immobilization of a protein of interest, or capture of the binding partners of a protein of interest within its biochemical environment. The HaloTag is composed of two covalently bound segments including a haloalkane dehalogenase and a synthetic ligand of choice. These synthetic ligands consist of a reactive chloroalkane linker bound to a functional group. Functional groups can either be biotin (can be used as an affinity tag) or can be chosen from five availa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescent Dye
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with several π bonds. Fluorophores are sometimes used alone, as a tracer in fluids, as a dye for staining of certain structures, as a substrate of enzymes, or as a probe or indicator (when its fluorescence is affected by environmental aspects such as polarity or ions). More generally they are covalently bonded to a macromolecule, serving as a marker (or dye, or tag, or reporter) for affine or bioactive reagents ( antibodies, peptides, nucleic acids). Fluorophores are notably used to stain tissues, cells, or materials in a variety of analytical methods, i.e., fluorescent imaging and spectroscopy. Fluorescein, via its amine-reactive isothiocyanate derivative fluorescein isothiocyanate (FITC), has been one of the most popular fluorophores. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biochemistry
Biochemistry or biological chemistry is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology and metabolism. Over the last decades of the 20th century, biochemistry has become successful at explaining living processes through these three disciplines. Almost all areas of the life sciences are being uncovered and developed through biochemical methodology and research. Voet (2005), p. 3. Biochemistry focuses on understanding the chemical basis which allows biological molecules to give rise to the processes that occur within living cells and between cells,Karp (2009), p. 2. in turn relating greatly to the understanding of tissues and organs, as well as organism structure and function.Miller (2012). p. 62. Biochemistry is closely related to molecular biology, which is the study of the molecular mechanisms of biological phenomena.As ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzymes
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are Ribozyme, catalytic RNA molecules, called ribozymes. Enzymes' Chemical specificity, specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Green Fluorescent Protein
The green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range. The label ''GFP'' traditionally refers to the protein first isolated from the jellyfish ''Aequorea victoria'' and is sometimes called ''avGFP''. However, GFPs have been found in other organisms including corals, sea anemones, zoanithids, copepods and lancelets. The GFP from ''A. victoria'' has a major excitation peak at a wavelength of 395 nm and a minor one at 475 nm. Its emission peak is at 509 nm, which is in the lower green portion of the visible spectrum. The fluorescence quantum yield (QY) of GFP is 0.79. The GFP from the sea pansy (''Renilla reniformis'') has a single major excitation peak at 498 nm. GFP makes for an excellent tool in many forms of biology due to its ability to form an internal chromophore without requiring any accessory cofactors, gene products, or enzymes / substrates other than mo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Yellow Fluorescent Protein
Yellow fluorescent protein (YFP) is a genetic mutant of green fluorescent protein (GFP) originally derived from the jellyfish ''Aequorea victoria''. Its excitation peak is 513 nm and its emission peak is 527 nm. Like the parent GFP, YFP is a useful tool in cell and molecular biology because the excitation and emission peaks of YFP are distinguishable from GFP which allows for the study of multiple processes/proteins within the same experiment. Three improved versions of YFP are Citrine, Venus, and Ypet. They have reduced chloride sensitivity, faster maturation, and increased brightness (defined as the product of the extinction coefficient and quantum yield). Typically, YFP serves as the acceptor for genetically-encoded FRET sensors of which the most likely donor FP is monomeric cyan fluorescent protein (mCFP). The red-shift relative to GFP is caused by a Pi-Pi stacking interaction as a result of the T203Y substitution introduced by mutation, which essentially increase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
O-6-methylguanine-DNA Methyltransferase
''O''6-alkylguanine DNA alkyltransferase (also known as AGT, MGMT or AGAT) is a protein that in humans is encoded by the ''O''6-methylguanine DNA methyltransferase (''MGMT'') gene. O6-methylguanine DNA methyltransferase is crucial for genome stability. It repairs the naturally occurring mutagenic DNA lesion O6-methylguanine back to guanine and prevents mismatch and errors during DNA replication and transcription. Accordingly, loss of ''MGMT'' increases the carcinogenic risk in mice after exposure to alkylating agents. The two bacterial isozymes are Ada and Ogt. Function and mechanism Although alkylating mutagens preferentially modify the guanine base at the N7 position, ''O''6-alkyl-guanine is a major carcinogenic lesion in DNA. This DNA adduct is removed by the repair protein ''O''6-alkylguanine DNA alkyltransferase through an SN2 mechanism. This protein is not a true enzyme since it removes the alkyl group from the lesion in a stoichiometric reaction and the active enzyme ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescence Microscopy
A fluorescence microscope is an optical microscope that uses fluorescence instead of, or in addition to, scattering, reflection, and attenuation or absorption, to study the properties of organic or inorganic substances. "Fluorescence microscope" refers to any microscope that uses fluorescence to generate an image, whether it is a simple set up like an epifluorescence microscope or a more complicated design such as a confocal microscope, which uses optical sectioning to get better resolution of the fluorescence image. Principle The specimen is illuminated with light of a specific wavelength (or wavelengths) which is absorbed by the fluorophores, causing them to emit light of longer wavelengths (i.e., of a different color than the absorbed light). The illumination light is separated from the much weaker emitted fluorescence through the use of a spectral emission filter. Typical components of a fluorescence microscope are a light source (xenon arc lamp or mercury-vapor lamp are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
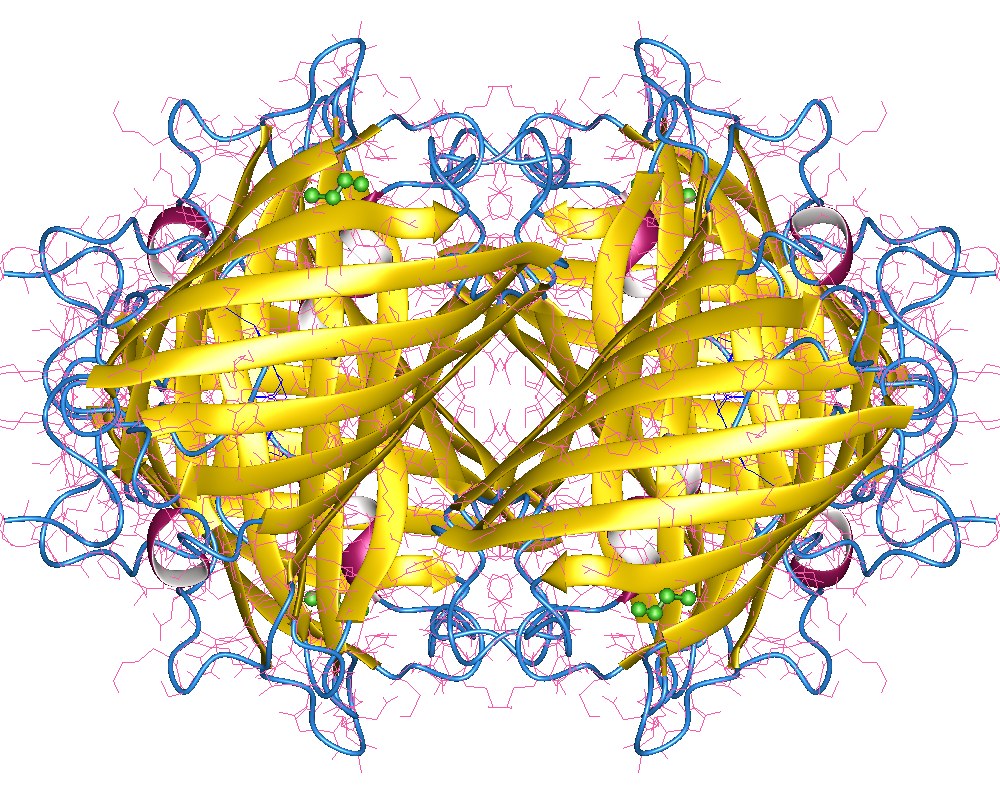
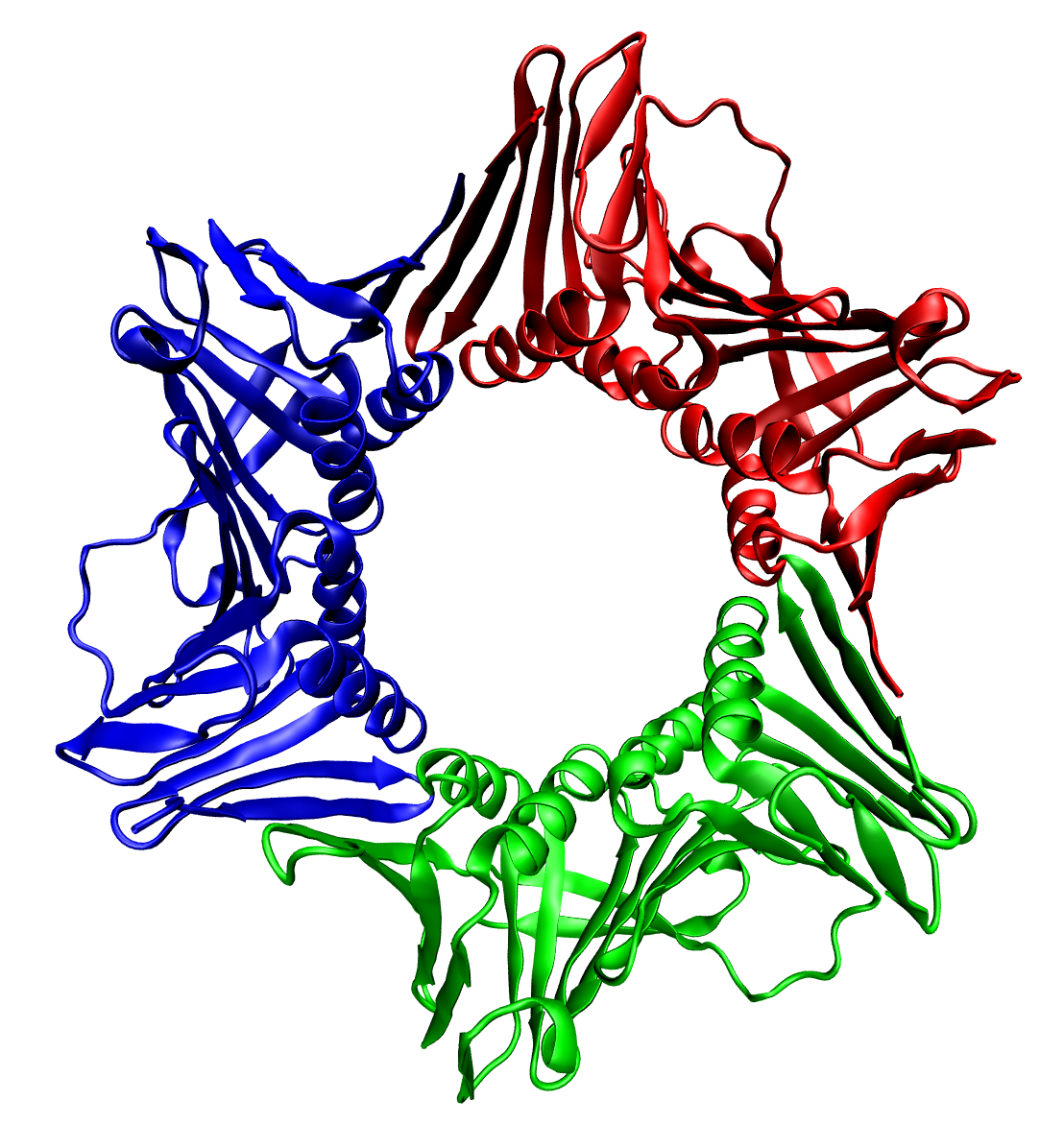
Multiprotein Complexes
Protein quaternary structure is the fourth (and highest) classification level of protein structure. Protein quaternary structure refers to the structure of proteins which are themselves composed of two or more smaller protein chains (also referred to as subunits). Protein quaternary structure describes the number and arrangement of multiple folded protein subunits in a multi-subunit complex. It includes organizations from simple dimers to large homooligomers and complexes with defined or variable numbers of subunits. In contrast to the first three levels of protein structure, not all proteins will have a quaternary structure since some proteins function as single units. Protein quaternary structure can also refer to biomolecular complexes of proteins with nucleic acids and other cofactors. Description and examples Many proteins are actually assemblies of multiple polypeptide chains. The quaternary structure refers to the number and arrangement of the protein subunits wit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |