|
SprD
__NOTOC__ In molecular biology SprD (Small pathogenicity island RNA D) is a non-coding RNA expressed on pathogenicity islands in ''Staphylococcus aureus''. It was identified ''in silico'' along with a number of other sRNAs (SprA-G) through microarray analysis which were confirmed using a Northern blot. SprD has been found to significantly contribute to causing disease in an animal model. Function SprD is located between genes ''scn'' and ''chp'' in the innate immune evasion cluster (IEC) of the ''S. aureus'' genome. Its placement within this region was the first indication of a virulence-factor regulatory function. SprD binds with ''sbi'' (''Staphylococcus aureus'' binder of IgG) mRNA which encodes an immune evasion protein. It occludes the Shine-Dalgarno sequence and the initiation codon of ''sbi'', forming a ''sbi'' mRNA-SprD duplex repressing the translation of the mRNA. ''sbi'' protein interferes with the host's innate immune response by binding Factor H, Complement compo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
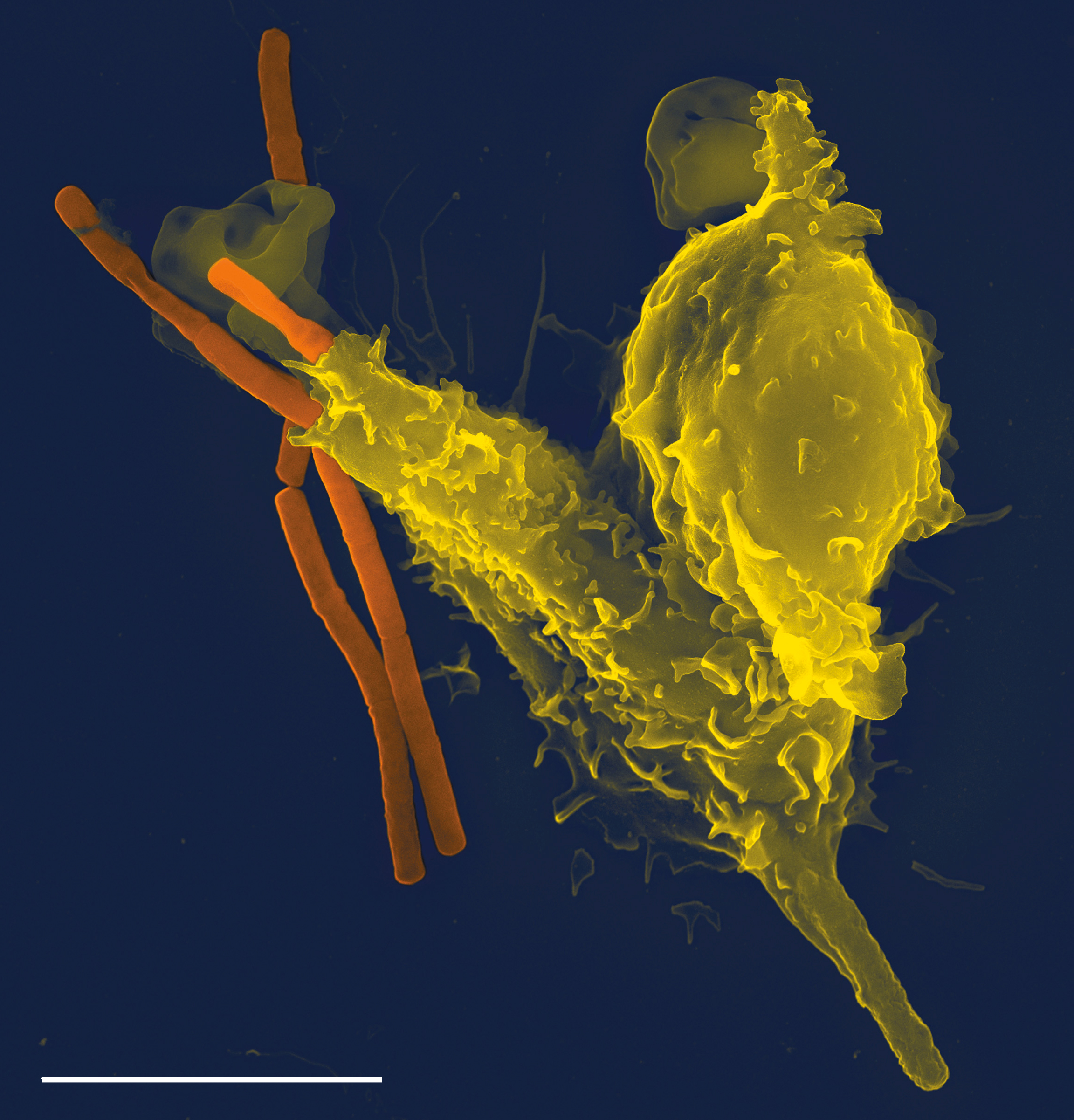
Staphylococcus Aureus
''Staphylococcus aureus'' is a Gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe that can grow without the need for oxygen. Although ''S. aureus'' usually acts as a commensal of the human microbiota, it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. ''S. aureus'' is one of the leading pathogens for deaths associated with antimicrobial resistance and the emergence of antibiotic-resistant strains, such as methicillin-resistant ''S. aureus'' (MRSA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SprX Small RNA
In molecular biology the small pathogenicity island RNA X (alias RsaOR) gene is a bacterial non-coding RNA. It was discovered in a large-scale analysis of ''Staphylococcus aureus''. SprX was shown to influence antibiotic resistance of the bacteria to Vancomycin and Teicoplanin glycopeptides, which are used to treat MRSA infections. In this study the authors identified a SprX target, stage V sporulation protein G (Spo VG). By reducing Spo VG expression levels, SprX affects ''S. aureus'' resistance to the glycopeptide antibiotics. Further work demonstrated its involvement in the regulation of pathogenicity factors. See also * SprD __NOTOC__ In molecular biology SprD (Small pathogenicity island RNA D) is a non-coding RNA expressed on pathogenicity islands in ''Staphylococcus aureus''. It was identified ''in silico'' along with a number of other sRNAs (SprA-G) through microar ... References Non-coding RNA {{molecular-cell-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR. The number of non-coding RNAs within the human genome is unknown; however, recent transcriptomic and bioinformatic studies suggest that there are thousands of non-coding transcripts. Many of the newly identified ncRNAs have not been validated for their function. There is no consensus in the literature on how much of non-coding transcription is functional. Some researchers have argued that many ncRNAs are non-functional (sometimes referred to as "junk RNA"), spurious transcriptions. Others, however, disagree, arguing instead that many ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein. mRNA is created during the process of Transcription (biology), transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as Translation (biology), translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genet ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein A
Protein A is a 42 kDa surface protein originally found in the cell wall of the bacteria ''Staphylococcus aureus''. It is encoded by the ''spa'' gene and its regulation is controlled by DNA topology, cellular osmolarity, and a two-component system called ArlS-ArlR. It has found use in biochemical research because of its ability to bind immunoglobulins. It is composed of five homologous Ig-binding domains that fold into a three-helix bundle. Each domain is able to bind proteins from many mammalian species, most notably IgGs. It binds the heavy chain within the Fc region of most immunoglobulins and also within the Fab region in the case of the human VH3 family. Through these interactions in serum, where IgG molecules are bound in the wrong orientation (in relation to normal antibody function), the bacteria disrupts opsonization and phagocytosis. History As a by-product of his work on type-specific staphylococcus antigens, Verwey reported in 1940 that a protein fraction prepared from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Complement Component 3
Complement component 3, often simply called C3, is a protein of the immune system. It plays a central role in the complement system and contributes to innate immunity. In humans it is encoded on chromosome 19 by a gene called ''C3''. Function C3 plays a central role in the activation of the complement system. Its activation is required for both classical and alternative complement activation pathways. People with C3 deficiency are susceptible to bacterial infection. One form of C3-convertase, also known as C4b2a, is formed by a heterodimer of activated forms of C4 and C2. It catalyzes the proteolytic cleavage of C3 into C3a and C3b, generated during activation through the classical pathway as well as the lectin pathway. C3a is an anaphylotoxin and the precursor of some cytokines such as ASP, and C3b serves as an opsonizing agent. Factor I can cleave C3b into C3c and C3d, the latter of which plays a role in enhancing B cell responses. In the alternative complement pathway, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Factor H
Factor H is a member of the regulators of complement activation family and is a complement control protein. It is a large (155 kilodaltons), soluble glycoprotein that circulates in human plasma (at typical concentrations of 200–300 micrograms per milliliter). Its principal function is to regulate the alternative pathway of the complement system, ensuring that the complement system is directed towards pathogens or other dangerous material and does not damage host tissue. Factor H regulates complement activation on self cells and surfaces by possessing both cofactor activity for the Factor I mediated C3b cleavage, and decay accelerating activity against the alternative pathway C3-convertase, C3bBb. Factor H exerts its protective action on self cells and self surfaces but not on the surfaces of bacteria or viruses. This is thought to be the result of Factor H having the ability to adopt conformations with lower or higher activities as a cofactor for C3 cleavage or decay accelerat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translation (genetics)
In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription of DNA to RNA in the cell's nucleus. The entire process is called gene expression. In translation, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later folds into an active protein and performs its functions in the cell. The ribosome facilitates decoding by inducing the binding of complementary tRNA anticodon sequences to mRNA codons. The tRNAs carry specific amino acids that are chained together into a polypeptide as the mRNA passes through and is "read" by the ribosome. Translation proceeds in three phases: # Initiation: The ribosome assembles around the target mRNA. The first tRNA is attached at the start codon. # Elongation: The last tRNA validated by the small ribosomal subunit (''accommodation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Initiation Codon
The start codon is the first codon of a messenger RNA (mRNA) transcript translated by a ribosome. The start codon always codes for methionine in eukaryotes and Archaea and a N-formylmethionine (fMet) in bacteria, mitochondria and plastids. The most common start codon is AUG (i.e., ATG in the corresponding DNA sequence). The start codon is often preceded by a 5' untranslated region (5' UTR). In prokaryotes this includes the ribosome binding site. Alternative start codons Alternative start codons are different from the standard AUG codon and are found in both prokaryotes (bacteria and archaea) and eukaryotes. Alternate start codons are still translated as Met when they are at the start of a protein (even if the codon encodes a different amino acid otherwise). This is because a separate transfer RNA (tRNA) is used for initiation. Eukaryotes Alternate start codons (non-AUG) are very rare in eukaryotic genomes. However, naturally occurring non-AUG start codons have b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Immune System
The immune system is a network of biological processes that protects an organism from diseases. It detects and responds to a wide variety of pathogens, from viruses to parasitic worms, as well as cancer cells and objects such as wood splinters, distinguishing them from the organism's own healthy tissue. Many species have two major subsystems of the immune system. The innate immune system provides a preconfigured response to broad groups of situations and stimuli. The adaptive immune system provides a tailored response to each stimulus by learning to recognize molecules it has previously encountered. Both use molecules and cells to perform their functions. Nearly all organisms have some kind of immune system. Bacteria have a rudimentary immune system in the form of enzymes that protect against virus infections. Other basic immune mechanisms evolved in ancient plants and animals and remain in their modern descendants. These mechanisms include phagocytosis, antimicrobial pe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virulence
Virulence is a pathogen's or microorganism's ability to cause damage to a host. In most, especially in animal systems, virulence refers to the degree of damage caused by a microbe to its host. The pathogenicity of an organism—its ability to cause disease—is determined by its virulence factors. In the specific context of gene for gene systems, often in plants, virulence refers to a pathogen's ability to infect a resistant host. The noun ''virulence'' derives from the adjective ''virulent'', meaning disease severity. The word ''virulent'' derives from the Latin word ''virulentus'', meaning "a poisoned wound" or "full of poison." From an ecological standpoint, virulence is the loss of fitness induced by a parasite upon its host. Virulence can be understood in terms of proximate causes—those specific traits of the pathogen that help make the host ill—and ultimate causes—the evolutionary pressures that lead to virulent traits occurring in a pathogen strain. Virulent ba ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |