|
Single-cell DNA Template Strand Sequencing
Single-cell DNA template strand sequencing, or Strand-seq, is a technique for the selective sequencing of a daughter cell's parental template strands. This technique offers a wide variety of applications, including the identification of sister chromatid exchanges in the parental cell prior to segregation, the assessment of non-random segregation of sister chromatids, the identification of misoriented contigs in genome assemblies, de novo genome assembly of both haplotypes in diploid organisms including humans, whole-chromosome haplotyping, and the identification of germline and somatic genomic structural variation, the latter of which can be detected robustly even in single cells. Background Strand-seq (single-cell and single-strand sequencing) was one of the first single-cell sequencing protocols described in 2012. This genomic technique selectively sequencings the parental template strands in single daughter cells DNA libraries. As a proof of concept study, the authors d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
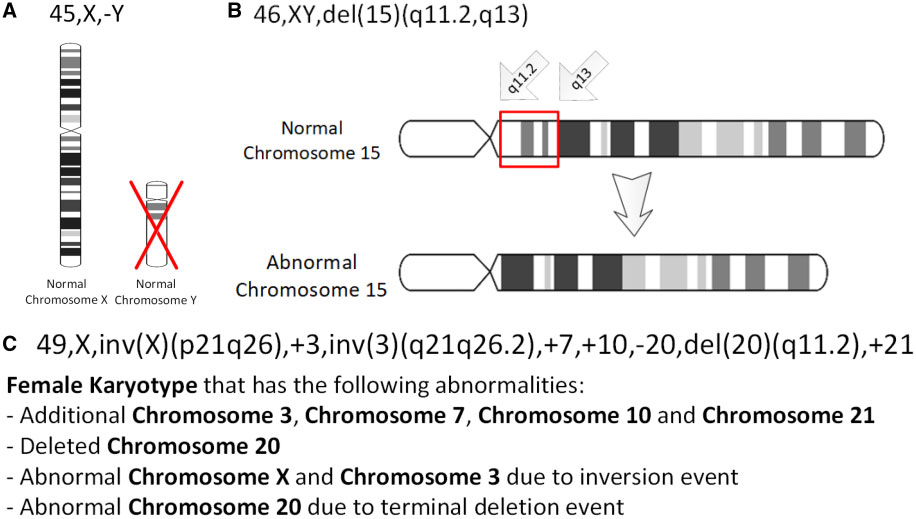
Chromosomal Inversion
An inversion is a chromosome rearrangement in which a segment of a chromosome becomes inverted within its original position. An inversion occurs when a chromosome undergoes a two breaks within the chromosomal arm, and the segment between the two breaks inserts itself in the opposite direction in the same chromosome arm. The breakpoints of inversions often happen in regions of repetitive nucleotides, and the regions may be reused in other inversions. Chromosomal segments in inversions can be as small as 100 kilobases or as large as 100 megabases. The number of genes captured by an inversion can range from a handful of genes to hundreds of genes. Inversions can happen either through ectopic recombination, chromosomal breakage and repair, or non-homologous end joining. Inversions are of two types: paracentric and pericentric. Paracentric inversions do not include the centromere, and both breakpoints occur in one arm of the chromosome. Pericentric inversions span the centromere, and t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aneuploidy
Aneuploidy is the presence of an abnormal number of chromosomes in a cell, for example a human cell having 45 or 47 chromosomes instead of the usual 46. It does not include a difference of one or more complete sets of chromosomes. A cell with any number of complete chromosome sets is called a '' euploid'' cell. An extra or missing chromosome is a common cause of some genetic disorders. Some cancer cells also have abnormal numbers of chromosomes. About 68% of human solid tumors are aneuploid. Aneuploidy originates during cell division when the chromosomes do not separate properly between the two cells ( nondisjunction). Most cases of aneuploidy in the autosomes result in miscarriage, and the most common extra autosomal chromosomes among live births are 21, 18 and 13. Chromosome abnormalities are detected in 1 of 160 live human births. Autosomal aneuploidy is more dangerous than sex chromosome aneuploidy, as autosomal aneuploidy is almost always lethal to embryos that cease ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Sequence Alignment Software
This list of sequence alignment software is a compilation of software tools and web portals used in pairwise sequence alignment and multiple sequence alignment. See structural alignment software for structural alignment of proteins. Database search only *Sequence type: protein or nucleotide Pairwise alignment *Sequence type: protein or nucleotide **Alignment type: local or global Multiple sequence alignment *Sequence type: protein or nucleotide. **Alignment type: local or global Genomics analysis *Sequence type: protein or nucleotide Motif finding *Sequence type: protein or nucleotide Benchmarking Alignment viewers, editors Please see List of alignment visualization software. Short-read sequence alignment See also * List of open source bioinformatics software References {{Reflist Genetics-related lists, Sequence Lists of bioinformatics software, Sequence alignment software ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatic Analysis Of Inherited Templates (BAIT) Output
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for ''in silico'' analyses of biological queries using computational and statistical techniques. Bioinformatics includes biological studies that use computer programming as part of their methodology, as well as specific analysis "pipelines" that are repeatedly used, particularly in the field of genomics. Common uses of bioinformatics include the identification of candidates genes and single nucleotide polymorphisms (SNPs). Often, such identification is made with the aim to better understand the genetic basis of disease, unique adaptations, desirable properties (es ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MALBAC
Multiple Annealing and Looping Based Amplification Cycles (MALBAC) is a quasilinear whole genome amplification method. Unlike conventional DNA amplification methods that are non-linear or exponential (in each cycle, DNA copied can serve as template for subsequent cycles), MALBAC utilizes special primers that allow amplicons to have complementary ends and therefore to loop, preventing DNA from being copied exponentially. This results in amplification of only the original genomic DNA and therefore reduces amplification bias. MALBAC is “used to create overlapped shotgun amplicons covering most of the genome”.Zong, C.; Lu, S.; Chapman, A.R.; Xie, S. (2012). "Genome-wide detection of single-nucleotide and copy-number variations of a single human cell." ''Science'' 338, 1622. DOI: 10.1126/science.1229164. For next generation sequencing, MALBAC is followed by regular PCR which is used to further amplify amplicons. Technological platform Prior to MALBAC, a single cell is isolated ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Multiple Displacement Amplification
Multiple displacement amplification (MDA) is a DNA amplification technique. This method can rapidly amplify minute amounts of DNA samples to a reasonable quantity for genomic analysis. The reaction starts by annealing random hexamer primers to the template: DNA synthesis is carried out by a high fidelity enzyme, preferentially Φ29 DNA polymerase. Compared with conventional PCR amplification techniques, MDA does not employ sequence-specific primers but amplifies all DNA, generates larger-sized products with a lower error frequency, and works at a constant temperature. MDA has been actively used in whole genome amplification (WGA) and is a promising method for application to single cell genome sequencing and sequencing-based genetic studies. Background Many biological and forensic cases involving genetic analysis require sequencing of DNA from minute amounts of sample, such as DNA from uncultured single cells or trace amounts of tissue collected from crime scenes. Conventional ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single Cell Sequencing
Single-cell sequencing examines the sequence information from individual cells with optimized next-generation sequencing technologies, providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment. For example, in cancer, sequencing the DNA of individual cells can give information about mutations carried by small populations of cells. In development, sequencing the RNAs expressed by individual cells can give insight into the existence and behavior of different cell types. In microbial systems, a population of the same species can appear genetically clonal. Still, single-cell sequencing of RNA or epigenetic modifications can reveal cell-to-cell variability that may help populations rapidly adapt to survive in changing environments. Background A typical human cell consists of about 2 x 3.3 billion base pairs of DNA and 600 million mRNA bases. Usually, a mix of millions of cells is used ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical research ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Illumina (company)
Illumina, Inc. is an American biotechnology company, headquartered in San Diego, California. Incorporated on April 1, 1998, Illumina develops, manufactures, and markets integrated systems for the analysis of genetic variation and biological function. The company provides a line of products and services that serves the sequencing, genotyping and gene expression, and proteomics markets. Illumina's technology had purportedly reduced the cost of sequencing a human genome to by 2014. Its customers include genomic research centers, pharmaceutical companies, academic institutions, clinical research organizations, and biotechnology companies. History Illumina was founded in April 1998 by David Walt, Larry Bock, John Stuelpnagel, Anthony Czarnik, and Mark Chee. While working with CW Group, a venture-capital firm, Bock and Stuelpnagel uncovered what would become Illumina's BeadArray technology at Tufts University and negotiated an exclusive license to that technology. In 1999, Illumin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lysis Buffer
A lysis buffer is a buffer solution used for the purpose of breaking open cells for use in molecular biology experiments that analyze the labile macromolecules of the cells (e.g. western blot for protein, or for DNA extraction). Most lysis buffers contain buffering salts (e.g. Tris-HCl) and ionic salts (e.g. NaCl) to regulate the pH and osmolarity of the lysate. Sometimes detergents (such as Triton X-100 or SDS) are added to break up membrane structures. For lysis buffers targeted at protein extraction, protease inhibitors are often included, and in difficult cases may be almost required. Lysis buffers can be used on both animal and plant tissue cells. Choosing a buffer The primary purpose of lysis buffer is isolating the molecules of interest and keeping them in a stable environment. For proteins, for some experiments, the target proteins should be completely denatured, while in some other experiments the target protein should remain folded and functional. Different prot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |





.jpg)