|
SRGAP2
SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2), also known as formin-binding protein 2 (FNBP2), is a mammalian protein that in humans is encoded by the ''SRGAP2'' gene. It is involved in neuronal migration and differentiation and plays a critical role in synaptic development, brain mass and number of cortical neurons. Downregulation of srGAP2 inhibits cell–cell repulsion and enhances cell–cell contact duration. SRGAP2 dimerizes through its F-BAR domain. SRGAP2C, a shortened version found in early hominins and humans that only has the F-BAR domain, antagonizes its action. It slows maturation of some neurons and increases neuronal spine density. Evolution ''SRGAP2'' is one of 23 genes that are known to be duplicated in humans but not other primates. ''SRGAP2'' has been duplicated three times in the human genome in the past 3.4 million years: one duplication 3.4 million years ago (mya) called SRGAP2B, followed by two that copied SRGAP2B 2.4 mya into SRGAP2C and ~1 mya in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SRGAP2C
SLIT-ROBO Rho GTPase activating protein 2C is a protein in humans that is encoded by the SRGAP2C gene. Cortical development gene Slit-Robo Rho GTPase-activating protein 2 ( SRGAP2) has been highly conserved over mammalian evolution, and human is the only lineage wherein gene duplications have occurred (three times). The promoter and first nine exons of SRGAP2 duplicated from 1q23.1 ( SRGAP2A) to 1q21.1 ( SRGAP2B) ∼3.4 million years ago (mya). Two larger duplications later copied SRGAP2B to chromosome 1p12 (SRGAP2C) and to proximal 1q21.1 ( SRGAP2D) ∼2.4 and ∼1 mya, respectively. Ancestral SRGAP2A and the derived SRGAP2C copy are fixed at diploid copy number two. In contrast, the SRGAP2B and SRGAP2D are highly copy number polymorphic, with normal individuals identified that completely lack these paralogscopies. SRGAP2C is the most likely duplicate to encode a functional protein and is among the most fixed human-specific duplicate genes. Incomplete duplication created a novel ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RhoGAP
RhoGAP domain is an evolutionary conserved protein domain of GTPase activating proteins towards Rho/ Rac/ Cdc42-like small GTPases. Human proteins containing this domain ABR; ARHGAP1; ARHGAP10; ARHGAP11A; ARHGAP11B; ARHGAP12; ARHGAP15; ARHGAP17; ARHGAP18; ARHGAP19; ARHGAP20; ARHGAP21; ARHGAP22; ARHGAP23; ARHGAP24; ARHGAP25; ARHGAP26; ARHGAP27; ARHGAP28; ARHGAP29; ARHGAP30; ARHGAP4; ARHGAP5; ARHGAP6; ARHGAP8; ARHGAP9; BCR; BPGAP1; C1; C5orf5; CDGAP; CENTD1; CENTD2; CENTD3; CHN1; CHN2; DEPDC1; DEPDC1A; DEPDC1B; DLC1; FAM13A1; FKSG42; GMIP; GRLF1; HMHA1; INPP5B; KIAA1688; LOC553158; MYO9A; MYO9B; OCRL; OPHN1; PIK3R1; PIK3R2; PRR5; RACGAP1; RACGAP1P; RALBP1; RICH2; RICS; SH3BP1; SLIT1; SNX26; SRGAP1; SRGAP2; SRGAP3; STARD13; STARD8 StAR-related lipid transfer domain protein 8 (STARD8) also known as d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mammalian
Mammals () are a group of vertebrate animals constituting the class Mammalia (), characterized by the presence of mammary glands which in females produce milk for feeding (nursing) their young, a neocortex (a region of the brain), fur or hair, and three middle ear bones. These characteristics distinguish them from reptiles (including birds) from which they diverged in the Carboniferous, over 300 million years ago. Around 6,400 extant species of mammals have been described divided into 29 orders. The largest orders, in terms of number of species, are the rodents, bats, and Eulipotyphla (hedgehogs, moles, shrews, and others). The next three are the Primates (including humans, apes, monkeys, and others), the Artiodactyla (cetaceans and even-toed ungulates), and the Carnivora (cats, dogs, seals, and others). In terms of cladistics, which reflects evolutionary history, mammals are the only living members of the Synapsida (synapsids); this clade, together with Sauropsida ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudogene
Pseudogenes are nonfunctional segments of DNA that resemble functional genes. Most arise as superfluous copies of functional genes, either directly by DNA duplication or indirectly by reverse transcription of an mRNA transcript. Pseudogenes are usually identified when genome sequence analysis finds gene-like sequences that lack regulatory sequences needed for transcription or translation, or whose coding sequences are obviously defective due to frameshifts or premature stop codons. Most non-bacterial genomes contain many pseudogenes, often as many as functional genes. This is not surprising, since various biological processes are expected to accidentally create pseudogenes, and there are no specialized mechanisms to remove them from genomes. Eventually pseudogenes may be deleted from their genomes by chance DNA replication or DNA repair errors, or they may accumulate so many mutational changes that they are no longer recognizable as former genes. Analysis of these degeneration ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
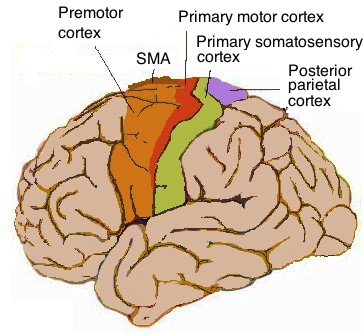
Cerebral Cortex
The cerebral cortex, also known as the cerebral mantle, is the outer layer of neural tissue of the cerebrum of the brain in humans and other mammals. The cerebral cortex mostly consists of the six-layered neocortex, with just 10% consisting of allocortex. It is separated into two cortices, by the longitudinal fissure that divides the cerebrum into the left and right cerebral hemispheres. The two hemispheres are joined beneath the cortex by the corpus callosum. The cerebral cortex is the largest site of neural integration in the central nervous system. It plays a key role in attention, perception, awareness, thought, memory, language, and consciousness. The cerebral cortex is part of the brain responsible for cognition. In most mammals, apart from small mammals that have small brains, the cerebral cortex is folded, providing a greater surface area in the confined volume of the cranium. Apart from minimising brain and cranial volume, cortical folding is crucial for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Filopodia
Filopodia (singular filopodium) are slender cytoplasmic projections that extend beyond the leading edge of lamellipodia in migrating cells. Within the lamellipodium, actin ribs are known as ''microspikes'', and when they extend beyond the lamellipodia, they're known as filopodia. They contain microfilaments (also called actin filaments) cross-linked into bundles by actin-bundling proteins, such as fascin and fimbrin. Filopodia form focal adhesions with the substratum, linking them to the cell surface. Many types of migrating cells display filopodia, which are thought to be involved in both sensation of chemotropic cues, and resulting changes in directed locomotion. Activation of the Rho family of GTPases, particularly cdc42 and their downstream intermediates, results in the polymerization of actin fibers by Ena/Vasp homology proteins. Growth factors bind to receptor tyrosine kinases resulting in the polymerization of actin filaments, which, when cross-linked, make up t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SH3 Domain
The SRC Homology 3 Domain (or SH3 domain) is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src. It has also been identified in several other protein families such as: PI3 Kinase, Ras GTPase-activating protein, CDC24 and cdc25. SH3 domains are found in proteins of signaling pathways regulating the cytoskeleton, the Ras protein, and the Src kinase and many others. The SH3 proteins interact with adaptor proteins and tyrosine kinases. Interacting with tyrosine kinases, SH3 proteins usually bind far away from the active site. Approximately 300 SH3 domains are found in proteins encoded in the human genome. In addition to that, the SH3 domain was responsible for controlling protein-protein interactions in the signal transduction pathways and regulating the intera ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
F-box
F-box proteins are proteins containing at least one F-box domain. The first identified F-box protein is one of three components of the SCF complex, which mediates ubiquitination of proteins targeted for degradation by the 26S proteasome. Core components F-box domain is a protein structural motif of about 50 amino acids that mediates protein–protein interactions. It has consensus sequence and varies in few positions. It was first identified in cyclin F. The F-box motif of Skp2, consisting of three alpha-helices, interacts directly with the SCF protein Skp1. F-box domains commonly exist in proteins in cancer with other protein–protein interaction motifs such as leucine-rich repeats (illustrated in the Figure) and WD repeats, which are thought to mediate interactions with SCF substrates. Function F-box proteins have also been associated with cellular functions such as signal transduction and regulation of the cell cycle. In plants, many F-box proteins are represented in gene ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neanderthal
Neanderthals (, also ''Homo neanderthalensis'' and erroneously ''Homo sapiens neanderthalensis''), also written as Neandertals, are an extinct species or subspecies of archaic humans who lived in Eurasia until about 40,000 years ago. While the "causes of Neanderthal disappearance about 40,000 years ago remain highly contested," demographic factors such as small population size, inbreeding and genetic drift, are considered probable factors. Other scholars have proposed competitive replacement, assimilation into the modern human genome (bred into extinction), great climatic change, disease, or a combination of these factors. It is unclear when the line of Neanderthals split from that of modern humans; studies have produced various intervals ranging from 315,000 to more than 800,000 years ago. The date of divergence of Neanderthals from their ancestor ''H. heidelbergensis'' is also unclear. The oldest potential Neanderthal bones date to 430,000 years ago, but the classificati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Denisovan
The Denisovans or Denisova hominins ) are an extinct species or subspecies of archaic human that ranged across Asia during the Lower and Middle Paleolithic. Denisovans are known from few physical remains and consequently, most of what is known about them comes from DNA evidence. No formal species name has been established pending more complete fossil material. The first identification of a Denisovan individual occurred in 2010, based on mitochondrial DNA (mtDNA) extracted from a juvenile female finger bone excavated from the Siberian Denisova Cave in the Altai Mountains in 2008. Nuclear DNA indicates close affinities with Neanderthals. The cave was also periodically inhabited by Neanderthals, but it is unclear whether Neanderthals and Denisovans ever cohabited in the cave. Additional specimens from Denisova Cave were subsequently identified, as was a single specimen from the Baishiya Karst Cave on the Tibetan Plateau. DNA evidence suggests they had dark skin, eyes, and hai ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Genome
The human genome is a complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondria. These are usually treated separately as the nuclear genome and the mitochondrial genome. Human genomes include both protein-coding DNA sequences and various types of DNA that does not encode proteins. The latter is a diverse category that includes DNA coding for non-translated RNA, such as that for ribosomal RNA, transfer RNA, ribozymes, small nuclear RNAs, and several types of regulatory RNAs. It also includes promoters and their associated gene-regulatory elements, DNA playing structural and replicatory roles, such as scaffolding regions, telomeres, centromeres, and origins of replication, plus large numbers of transposable elements, inserted viral DNA, non-functional pseudogenes and simple, highly-repetitive sequences. Introns make up a large percentage of non-coding DN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |