|
SEP15
15 kDa selenoprotein is a protein that in humans is encoded by the ''SEP15'' gene. Two alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. Function This gene encodes a selenoprotein, which contains a selenocysteine (Sec) residue at its active site. The selenocysteine is encoded by the UGA codon that normally signals translation termination. The 3' UTR of selenoprotein genes have a common stem-loop structure, the sec insertion sequence (SECIS), that is necessary for the recognition of UGA as a Sec codon rather than as a stop signal. Studies in mouse suggest that this selenoprotein may have redox function and may be involved in the quality control of protein folding. Clinical significance This gene is localized on chromosome 1p31, a genetic locus commonly mutated or deleted in human cancers. Protein domain The protein this gene encodes for is often called Sep15 however in the case of mice, it is named SelM. This protein is a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Selenoprotein
In molecular biology a selenoprotein is any protein that includes a selenocysteine (Sec, U, Se-Cys) amino acid residue. Among functionally characterized selenoproteins are five glutathione peroxidases (GPX) and three thioredoxin reductases, (TrxR/TXNRD) which both contain only one Sec. Selenoprotein P is the most common selenoprotein found in the plasma. It is unusual because in humans it contains 10 Sec residues, which are split into two domains, a longer N-terminal domain that contains 1 Sec, and a shorter C-terminal domain that contains 9 Sec. The longer N-terminal domain is likely an enzymatic domain, and the shorter C-terminal domain is likely a means of safely transporting the very reactive selenium atom throughout the body. Species distribution Selenoproteins exist in all major domains of life, eukaryotes, bacteria and archaea. Among eukaryotes, selenoproteins appear to be common in animals, but rare or absent in other phyla -one has been identified in the green alga ''C ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thioredoxin
Thioredoxin is a class of small redox proteins known to be present in all organisms. It plays a role in many important biological processes, including redox signaling. In humans, thioredoxins are encoded by ''TXN'' and ''TXN2'' genes. Loss-of-function mutation of either of the two human thioredoxin genes is lethal at the four-cell stage of the developing embryo. Although not entirely understood, thioredoxin is linked to medicine through their response to reactive oxygen species (ROS). In plants, thioredoxins regulate a spectrum of critical functions, ranging from photosynthesis to growth, flowering and the development and germination of seeds. Thioredoxins play a role in cell-to-cell communication. Occurrence They are found in nearly all known organisms and are essential for life in mammals. Function The primary function of Thioredoxin (Trx) is the reduction of oxidized cysteine residues and the cleavage of disulfide bonds. Multiple in vitro substrates for thioredoxin have be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Silencing
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene. Gene silencing can occur during either transcription or translation and is often used in research. In particular, methods used to silence genes are being increasingly used to produce therapeutics to combat cancer and other diseases, such as infectious diseases and neurodegenerative disorders. Gene silencing is often considered the same as gene knockdown. When genes are silenced, their expression is reduced. In contrast, when genes are knocked out, they are completely erased from the organism's genome and, thus, have no expression. Gene silencing is considered a gene knockdown mechanism since the methods used to silence genes, such as RNAi, CRISPR, or siRNA, generally reduce the expression of a gene by at least 70% but do not eliminate it. Methods using gene silencing are often considered better than gene knockouts since they allow researchers to study essential genes that are r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
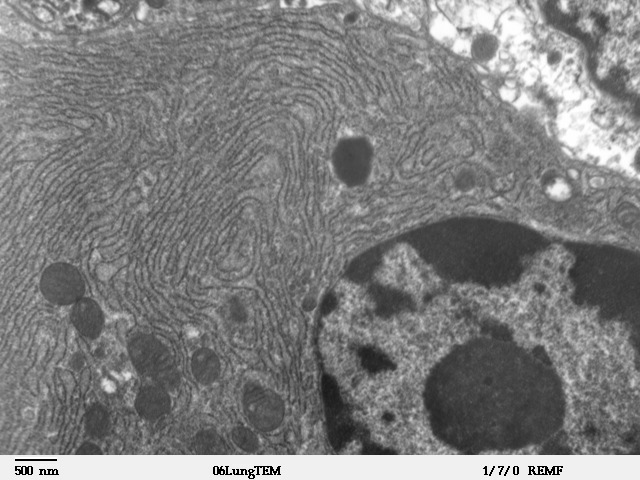
Endoplasmic Reticulum
The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum (RER), and smooth endoplasmic reticulum (SER). The endoplasmic reticulum is found in most eukaryotic cells and forms an interconnected network of flattened, membrane-enclosed sacs known as cisternae (in the RER), and tubular structures in the SER. The membranes of the ER are continuous with the outer nuclear membrane. The endoplasmic reticulum is not found in red blood cells, or spermatozoa. The two types of ER share many of the same proteins and engage in certain common activities such as the synthesis of certain lipids and cholesterol. Different types of cells contain different ratios of the two types of ER depending on the activities of the cell. RER is found mainly toward the nucleus of cell and SER towards the cell membrane or plasma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Bond
A chemical bond is a lasting attraction between atoms or ions that enables the formation of molecules and crystals. The bond may result from the electrostatic force between oppositely charged ions as in ionic bonds, or through the sharing of electrons as in covalent bonds. The strength of chemical bonds varies considerably; there are "strong bonds" or "primary bonds" such as covalent, ionic and metallic bonds, and "weak bonds" or "secondary bonds" such as dipole–dipole interactions, the London dispersion force and hydrogen bonding. Strong chemical bonding arises from the sharing or transfer of electrons between the participating atoms. Since opposite electric charges attract, the negatively charged electrons surrounding the nucleus and the positively charged protons within a nucleus attract each other. An electron positioned between two nuclei will be attracted to both of them, and the nuclei will be attracted toward electrons in this position. This attraction constitu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isomerase
Isomerases are a general class of enzymes that convert a molecule from one isomer to another. Isomerases facilitate intramolecular rearrangements in which bonds are broken and formed. The general form of such a reaction is as follows: A–B → B–A There is only one substrate yielding one product. This product has the same molecular formula as the substrate but differs in bond connectivity or spatial arrangement. Isomerases catalyze reactions across many biological processes, such as in glycolysis and carbohydrate metabolism. Isomerization Isomerases catalyze changes within one molecule. They convert one isomer to another, meaning that the end product has the same molecular formula but a different physical structure. Isomers themselves exist in many varieties but can generally be classified as structural isomers or stereoisomers. Structural isomers have a different ordering of bonds and/or different bond connectivity from one another, as in the case of hexane and it ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disulfide
In biochemistry, a disulfide (or disulphide in British English) refers to a functional group with the structure . The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. ''Persulfide'' usually refers to compounds. In inorganic chemistry disulfide usually refers to the corresponding anion (−S−S−). Organic disulfides Symmetrical disulfides are compounds of the formula . Most disulfides encountered in organo sulfur chemistry are symmetrical disulfides. Unsymmetrical disulfides (also called heterodisulfides) are compounds of the formula . They are less common in organic chemistry, but most disulfides in nature are unsymmetrical. Properties The disulfide bonds are strong, with a typical bond dissociation energy of 60 kcal/mol (251&nbs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Motif
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA. In nucleic acids Depending upon the sequence and other conditions, nucleic acids can form a variety of structural motifs which is thought to have biological significance. ;Stem-loop: Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Active Site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) and residues that catalyse a reaction of that substrate (catalytic site). Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes. Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity. This specificity is determined by the arrangement of amino acids within the active site and the structure of the substrates. Sometimes enzymes also need to bind with some cofactors to fulfil their function. The active si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Domain (biology)
In biological taxonomy, a domain ( or ) (Latin: ''regio''), also dominion, superkingdom, realm, or empire, is the highest taxonomic rank of all organisms taken together. It was introduced in the three-domain system of taxonomy devised by Carl Woese, Otto Kandler and Mark Wheelis in 1990. According to the domain system, the tree of life consists of either three domains such as Archaea, Bacteria, and Eukarya, or two domains consisting of Archaea and Bacteria, with Eukarya included in Archaea. The first two are all prokaryotes, single-celled microorganisms without a membrane-bound nucleus. All organisms that have a cell nucleus and other membrane-bound organelles are included in Eukarya. Non-cellular life is not included in this system. Alternatives to the three-domain system include the earlier two-empire system (with the empires Prokaryota and Eukaryota), and the eocyte hypothesis (with two domains of Bacteria and Archaea, with Eukarya included as a branch of Archaea). Term ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |