|
Regulation Of Transcription In Cancer
Generally, in progression to cancer, hundreds of genes are silenced or activated. Although silencing of some genes in cancers occurs by mutation, a large proportion of carcinogenic gene silencing is a result of altered DNA methylation (see DNA methylation in cancer). DNA methylation causing silencing in cancer typically occurs at multiple CpG sites in the CpG islands that are present in the promoters of protein coding genes. Altered expressions of microRNAs also silence or activate many genes in progression to cancer (see microRNAs in cancer). Altered microRNA expression occurs through hyper/hypo-methylation of CpG sites in CpG islands in promoters controlling transcription of the microRNAs. Silencing of DNA repair genes through methylation of CpG islands in their promoters appears to be especially important in progression to cancer (see methylation of DNA repair genes in cancer). CpG islands in promoters In humans, about 70% of promoters located near the transcription start ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Methylation In Cancer
DNA methylation in cancer plays a variety of roles, helping to change the healthy cells by regulation of gene expression to a cancer cells or a diseased cells disease pattern. One of the most widely studied DNA methylation dysregulation is the promoter hypermethylation where the CPGs islands in the promoter regions are methylated contributing or causing genes to be silenced. All mammalian cells descended from a fertilized egg (a zygote) share a common DNA sequence (except for new mutations in some lineages). However, during development and formation of different tissues epigenetic factors change. The changes include histone modifications, CpG island methylations and chromatin reorganizations which can cause the stable silencing or activation of particular genes. Once differentiated tissues are formed, CpG island methylation is generally stably inherited from one cell division to the next through the DNA methylation maintenance machinery. In cancer, a number of mutational change ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
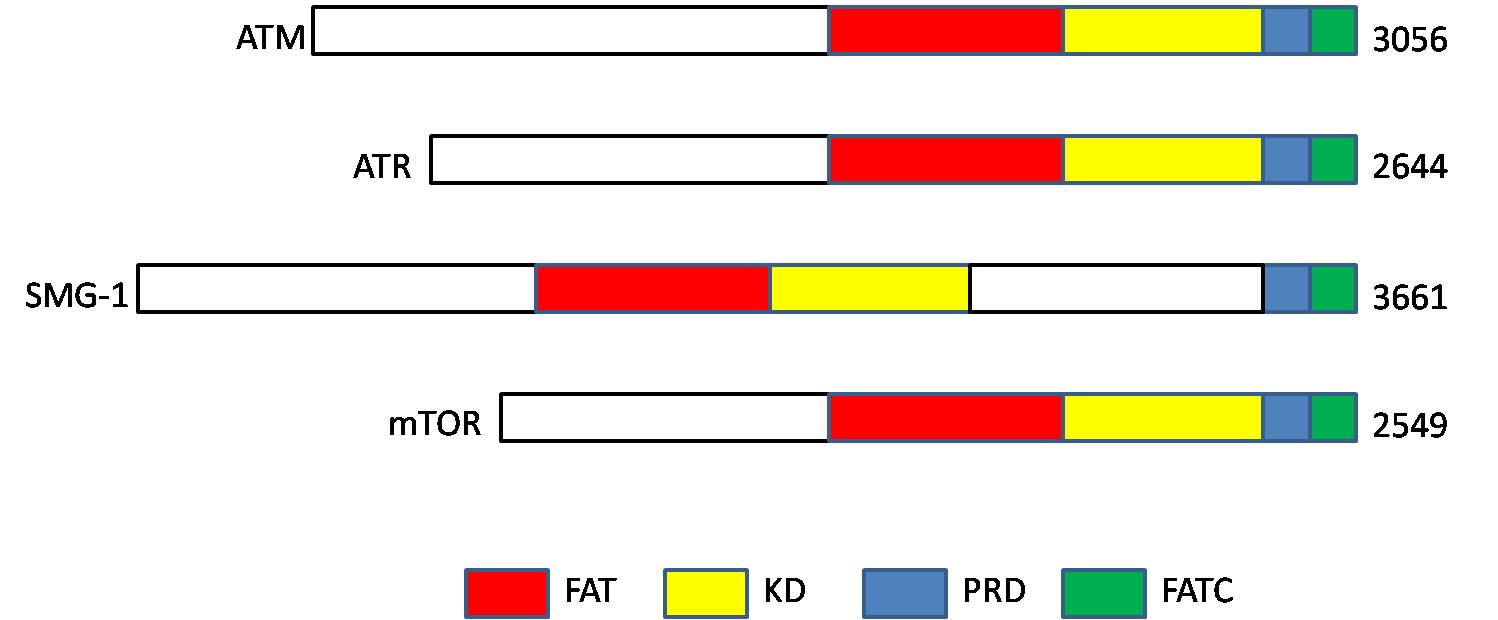
Ataxia Telangiectasia Mutated
ATM serine/threonine kinase or Ataxia-telangiectasia mutated, symbol ATM, is a serine/threonine protein kinase that is recruited and activated by DNA double-strand breaks. It phosphorylates several key proteins that initiate activation of the DNA damage checkpoint, leading to cell cycle arrest, DNA repair or apoptosis. Several of these targets, including p53, CHK2, BRCA1, NBS1 and H2AX are tumor suppressors. In 1995, the gene was discovered by Yosef Shiloh who named its product ATM since he found that its mutations are responsible for the disorder ataxia–telangiectasia#Cause, ataxia–telangiectasia. In 1998, the Shiloh and Michael B. Kastan, Kastan laboratories independently showed that ATM is a protein kinase whose activity is enhanced by DNA damage. Introduction Throughout the cell cycle DNA is monitored for damage. Damages result from errors during DNA replication, replication, by-products of metabolism, general toxic drugs or ionizing radiation. The cell cycle has diffe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-small-cell Lung Carcinoma
Non-small-cell lung cancer (NSCLC) is any type of epithelial lung cancer other than small-cell lung carcinoma (SCLC). NSCLC accounts for about 85% of all lung cancers. As a class, NSCLCs are relatively insensitive to chemotherapy, compared to small-cell carcinoma. When possible, they are primarily treated by surgical resection with curative intent, although chemotherapy has been used increasingly both preoperatively (Neoadjuvant therapy, neoadjuvant chemotherapy) and postoperatively (Adjuvant therapy, adjuvant chemotherapy). Types The most common types of NSCLC are squamous-cell carcinoma, large-cell carcinoma, and adenocarcinoma, but several other types occur less frequently. A few of the less common types are pleomorphic, carcinoid tumor, salivary gland carcinoma, and unclassified carcinoma. All types can occur in unusual histologic variants and as mixed cell-type combinations. Nonsquamous-cell carcinoma almost occupies the half of NSCLC. In the tissue classification, the cent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Head And Neck Cancer
Head and neck cancer develops from tissues in the lip and oral cavity (mouth), larynx (throat), salivary glands, nose, sinuses or the skin of the face. The most common types of head and neck cancers occur in the lip, mouth, and larynx. Symptoms predominantly include a sore that does not heal or a change in the voice. Some may experience a sore throat that does not go away. In those with advanced disease, there may be unusual bleeding, facial pain, numbness or swelling, and visible lumps on the outside of the neck or oral cavity. Given the location of these cancers, trouble breathing may also be present. The majority of head and neck cancer is caused by the use of alcohol or tobacco, including smokeless tobacco, with increasing cases linked to the human papillomavirus (HPV). Other risk factors include Epstein-Barr virus, betel quid, radiation exposure, certain workplace exposures. About 90% are pathologically classified as squamous cell cancers. The diagnosis is confirmed by t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NEIL1
Endonuclease VIII-like 1 is an enzyme that in humans is encoded by the ''NEIL1'' gene. NEIL1 belongs to a class of DNA glycosylases homologous to the bacterial Fpg/Nei family. These glycosylases initiate the first step in base excision repair by cleaving bases damaged by reactive oxygen species (ROS) and introducing a DNA strand break via the associated lyase reaction. Targets NEIL1 recognizes (targets) and removes certain ROS-damaged bases and then incises the abasic site via β,δ elimination, leaving 3′ and 5′ phosphate ends. NEIL1 recognizes oxidized pyrimidines, formamidopyrimidines, thymine residues oxidized at the methyl group, and both stereoisomers of thymine glycol. The best substrates for human NEIL1 appear to be the hydantoin lesions, guanidinohydantoin, and spiroiminodihydantoin that are further oxidation products of 8-oxoG. NEIL1 is also capable of removing lesions from single-stranded DNA as well as from bubble and forked DNA structures. Because the expre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LIG4
DNA ligase 4 is an enzyme that in humans is encoded by the LIG4 gene. Function The protein encoded by this gene is an ATP-dependent DNA ligase that joins double-strand breaks during the non-homologous end joining pathway of double-strand break repair. It is also essential for V(D)J recombination. Lig4 forms a complex with XRCC4, and further interacts with the DNA-dependent protein kinase (DNA-PK) and XLF/Cernunnos, which are also required for NHEJ. The crystal structure of the Lig4/XRCC4 complex has been resolved. Defects in this gene are the cause of LIG4 syndrome. The yeast homolog of Lig4 is Dnl4. LIG4 Syndrome In humans, deficiency of DNA ligase 4 results in a clinical condition known as LIG4 syndrome. This syndrome is characterized by cellular radiation sensitivity, growth retardation, developmental delay, microcephaly, facial dysmorphisms, increased disposition to leukemia, variable degrees of immunodeficiency and reduced number of blood cells. Haematopoietic st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
O-6-methylguanine-DNA Methyltransferase
''O''6-alkylguanine DNA alkyltransferase (also known as AGT, MGMT or AGAT) is a protein that in humans is encoded by the ''O''6-methylguanine DNA methyltransferase (''MGMT'') gene. O6-methylguanine DNA methyltransferase is crucial for genome stability. It repairs the naturally occurring mutagenic DNA lesion O6-methylguanine back to guanine and prevents mismatch and errors during DNA replication and transcription. Accordingly, loss of ''MGMT'' increases the carcinogenic risk in mice after exposure to alkylating agents. The two bacterial isozymes are Ada and Ogt. Function and mechanism Although alkylating mutagens preferentially modify the guanine base at the N7 position, ''O''6-alkyl-guanine is a major carcinogenic lesion in DNA. This DNA adduct is removed by the repair protein ''O''6-alkylguanine DNA alkyltransferase through an SN2 mechanism. This protein is not a true enzyme since it removes the alkyl group from the lesion in a stoichiometric reaction and the active enzyme ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Head And Neck Cancer
Head and neck cancer develops from tissues in the lip and oral cavity (mouth), larynx (throat), salivary glands, nose, sinuses or the skin of the face. The most common types of head and neck cancers occur in the lip, mouth, and larynx. Symptoms predominantly include a sore that does not heal or a change in the voice. Some may experience a sore throat that does not go away. In those with advanced disease, there may be unusual bleeding, facial pain, numbness or swelling, and visible lumps on the outside of the neck or oral cavity. Given the location of these cancers, trouble breathing may also be present. The majority of head and neck cancer is caused by the use of alcohol or tobacco, including smokeless tobacco, with increasing cases linked to the human papillomavirus (HPV). Other risk factors include Epstein-Barr virus, betel quid, radiation exposure, certain workplace exposures. About 90% are pathologically classified as squamous cell cancers. The diagnosis is confirmed by t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Post-transcriptional Regulation
Post-transcriptional regulation is the control of gene expression at the RNA level. It occurs once the RNA polymerase has been attached to the gene's promoter and is synthesizing the nucleotide sequence. Therefore, as the name indicates, it occurs between the transcription phase and the translation phase of gene expression. These controls are critical for the regulation of many genes across human tissues. It also plays a big role in cell physiology, being implicated in pathologies such as cancer and neurodegenerative diseases. Mechanism After being produced, the stability and distribution of the different transcripts is regulated (post-transcriptional regulation) by means of RNA binding protein (RBP) that control the various steps and rates controlling events such as alternative splicing, nuclear degradation ( exosome), processing, nuclear export (three alternative pathways), sequestration in P-bodies for storage or degradation and ultimately translation. These proteins ac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, genetic inf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Set Enrichment
Gene set enrichment analysis (GSEA) (also called functional enrichment analysis or pathway enrichment analysis) is a method to identify classes of genes or proteins that are over-represented in a large set of genes or proteins, and may have an association with disease phenotypes. The method uses statistical approaches to identify significantly enriched or depleted groups of genes. Transcriptomics technologies and proteomics results often identify thousands of genes which are used for the analysis. Researchers performing high-throughput experiments that yield sets of genes (for example, genes that are differentially expressed under different conditions) often want to retrieve a functional profile of that gene set, in order to better understand the underlying biological processes. This can be done by comparing the input gene set to each of the bins (terms) in the gene ontology – a statistical test can be performed for each bin to see if it is enriched for the input genes. Ba ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
_squamous_cell_carcinoma_histopathology.jpg)
