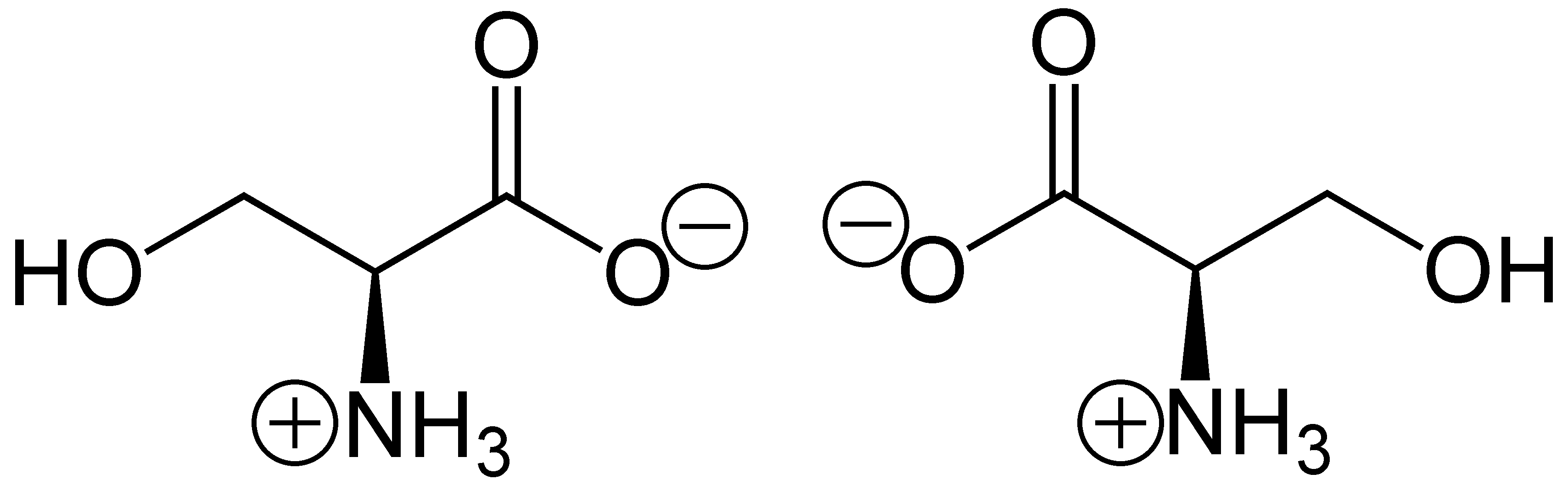|
Ataxia Telangiectasia Mutated
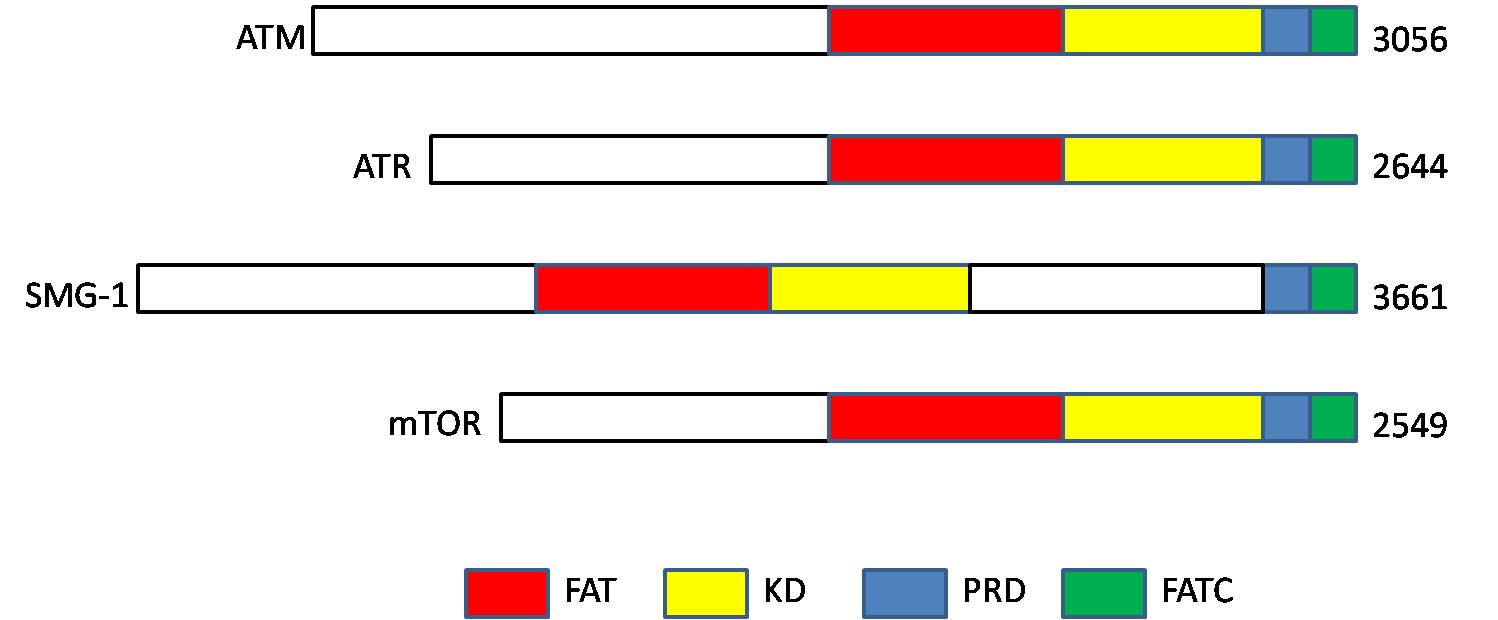
ATM serine/threonine kinase or Ataxia-telangiectasia mutated, symbol ATM, is a serine/threonine protein kinase that is recruited and activated by DNA double-strand breaks. It phosphorylates several key proteins that initiate activation of the DNA damage checkpoint, leading to cell cycle arrest, DNA repair or apoptosis. Several of these targets, including p53, CHK2, BRCA1, NBS1 and H2AX are tumor suppressors. In 1995, the gene was discovered by Yosef Shiloh who named its product ATM since he found that its mutations are responsible for the disorder ataxia–telangiectasia#Cause, ataxia–telangiectasia. In 1998, the Shiloh and Michael B. Kastan, Kastan laboratories independently showed that ATM is a protein kinase whose activity is enhanced by DNA damage. Introduction Throughout the cell cycle DNA is monitored for damage. Damages result from errors during DNA replication, replication, by-products of metabolism, general toxic drugs or ionizing radiation. The cell cycle has diffe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L-stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure was estab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Double-strand Breaks
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages (interstrand crosslinks or ICLs). This can eventually lead to malignant t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MRE11
Double-strand break repair protein MRE11 is an enzyme that in humans is encoded by the ''MRE11'' gene. The gene has been designated ''MRE11A'' to distinguish it from the pseudogene ''MRE11B'' that is nowadays named ''MRE11P1''. Function This gene encodes a nuclear protein involved in homologous recombination, telomere length maintenance, and DNA double-strand break repair. By itself, the protein has 3' to 5' exonuclease activity and endonuclease activity. The protein forms a complex with the RAD50 homolog; this complex is required for nonhomologous joining of DNA ends and possesses increased single-stranded DNA endonuclease and 3' to 5' exonuclease activities. In conjunction with a DNA ligase, this protein promotes the joining of noncomplementary ends in vitro using short homologies near the ends of the DNA fragments. This gene has a pseudogene on chromosome 3. Alternative splicing of this gene results in two transcript variants encoding different isoforms. Orthologs Mre11, an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disulfide Bond
In biochemistry, a disulfide (or disulphide in British English) refers to a functional group with the structure . The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. ''Persulfide'' usually refers to compounds. In inorganic chemistry disulfide usually refers to the corresponding anion (−S−S−). Organic disulfides Symmetrical disulfides are compounds of the formula . Most disulfides encountered in organo sulfur chemistry are symmetrical disulfides. Unsymmetrical disulfides (also called heterodisulfides) are compounds of the formula . They are less common in organic chemistry, but most disulfides in nature are unsymmetrical. Properties The disulfide bonds are strong, with a typical bond dissociation energy of 60 kcal/mol (251& ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
α-helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Huntingtin
Huntingtin (Htt) is the protein coded for in humans by the ''HTT'' gene, also known as the ''IT15'' ("interesting transcript 15") gene. Mutated ''HTT'' is the cause of Huntington's disease (HD), and has been investigated for this role and also for its involvement in long-term memory storage. It is variable in its structure, as the many polymorphisms of the gene can lead to variable numbers of glutamine residues present in the protein. In its wild-type (normal form), it contains 6-35 glutamine residues. However, in individuals affected by Huntington's disease (an autosomal dominant genetic disorder), it contains more than 36 glutamine residues (highest reported repeat length is about 250). Its commonly used name is derived from this disease; previously, the ''IT15'' label was commonly used. The mass of huntingtin protein is dependent largely on the number of glutamine residues it has; the predicted mass is around 350 kDa. Normal huntingtin is generally accepted to be 3144 am ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tertiary Structure
Protein tertiary structure is the three dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure.Branden C. and Tooze J. "Introduction to Protein Structure" Garland Publishing, New York. 1990 and 1991. A number of tertiary structures may fold into a quaternary structure.Kyte, J. "Structure in Protein Chemistry." Garland Publishing, New York. 1995. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of polypept ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transformation/transcription Domain-associated Protein
Transformation/transcription domain-associated protein, also known as TRRAP, is a protein that in humans is encoded by the ''TRRAP'' gene. TRRAP belongs to the phosphatidylinositol 3-kinase-related kinase protein family. Function TRRAP is an adaptor protein, which is found in various multiprotein chromatin complexes with histone acetyltransferase activity (HAT), which in turn is responsible for epigenetic transcription activation. TRRAP has a central role in MYC (c-Myc) transcription activation, and also participates in cell transformation by MYC. It is required for p53/TP53-, E2F1-, and E2F4-mediated transcription activation. It is also involved in transcription activation mediated by the adenovirus E1A, a viral oncoprotein that deregulates transcription of key genes. TRRAP is also required for the mitotic checkpoint and normal cell cycle progression. The MRN complex (composed of MRE11 Double-strand break repair protein MRE11 is an enzyme that in humans is encoded by the ''M ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HEAT Repeat Domain
A HEAT repeat is a protein tandem repeat structural motif composed of two alpha helices linked by a short loop. HEAT repeats can form alpha solenoids, a type of solenoid protein domain found in a number of cytoplasmic proteins. The name "HEAT" is an acronym for four proteins in which this repeat structure is found: Huntingtin, elongation factor 3 (EF3), protein phosphatase 2A (PP2A), and the yeast kinase TOR1. HEAT repeats form extended superhelical structures which are often involved in intracellular transport; they are structurally related to armadillo repeats. The nuclear transport protein importin beta contains 19 HEAT repeats. Various HEAT repeat proteins and their structures Representative examples of HEAT repeat proteins include importin β (also known as karyopherin β) family, regulatory subunits of condensin and cohesin, separase, PIKKs (phosphatidylinositol 3-kinase-related protein kinases) such as ATM ( Ataxia telangiectasia mutated) and ATR (Ataxia telangiectasia an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MTOR
The mammalian target of sirolimus, rapamycin (mTOR), also referred to as the mechanistic target of rapamycin, and sometimes called FK506-binding protein 12-rapamycin-associated protein 1 (FRAP1), is a kinase that in humans is encoded by the ''MTOR'' gene. mTOR is a member of the phosphatidylinositol 3-kinase-related kinase family of protein kinases. mTOR links with other proteins and serves as a core component of two distinct protein complexes, mTORC1, mTOR complex 1 and mTORC2, mTOR complex 2, which regulate different cellular processes. In particular, as a core component of both complexes, mTOR functions as a serine/threonine protein kinase that regulates cell growth, cell proliferation, cell motility, cell survival, protein synthesis, autophagy, and Transcription (genetics), transcription. As a core component of mTORC2, mTOR also functions as a tyrosine protein kinase that promotes the activation of insulin receptors and insulin-like growth factor 1 receptors. mTORC2 has also ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA-PKcs
DNA-dependent protein kinase, catalytic subunit, also known as DNA-PKcs, is an enzyme that in humans is encoded by the gene designated as ''PRKDC'' or ''XRCC7''. DNA-PKcs belongs to the phosphatidylinositol 3-kinase-related kinase protein family. The DNA-Pkcs protein is a serine/threonine protein kinase comprising a single polypeptide chain of 4,128 amino acids. Function DNA-PKcs is the catalytic subunit of a nuclear DNA-dependent serine/threonine protein kinase called DNA-PK. The second component is the autoimmune antigen Ku. On its own, DNA-PKcs is inactive and relies on Ku to direct it to DNA ends and trigger its kinase activity. DNA-PKcs is required for the non-homologous end joining (NHEJ) pathway of DNA repair, which rejoins double-strand breaks. It is also required for V(D)J recombination, a process that utilizes NHEJ to promote immune system diversity. DNA-PKcs knockout mice have severe combined immunodeficiency due to their V(D)J recombination defect. Many proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |