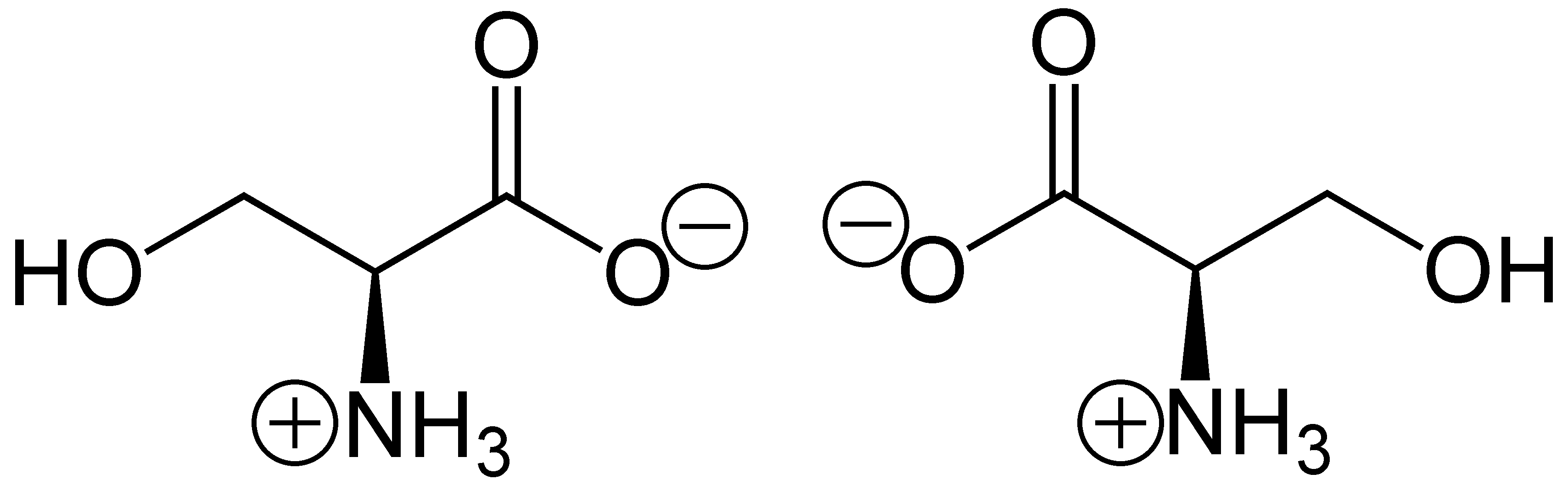|
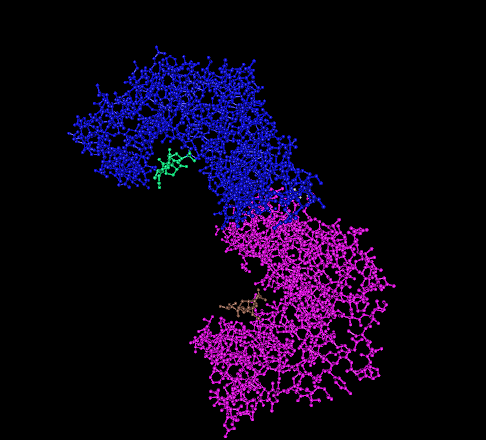
RAF-10
RAF proto-oncogene serine/threonine-protein kinase, also known as proto-oncogene c-RAF or simply c-Raf or even Raf-1, is an enzyme that in humans is encoded by the ''RAF1'' gene. The c-Raf protein is part of the ERK1/2 pathway as a MAP kinase (MAP3K) that functions downstream of the Ras subfamily of membrane associated GTPases. C-Raf is a member of the Raf kinase family of serine/threonine-specific protein kinases, from the TKL (Tyrosine-kinase-like) group of kinases. Discovery The first Raf gene, v-Raf was found in 1983. It was isolated from the murine retrovirus bearing the number 3611. It was soon demonstrated to be capable to transform rodent fibroblasts to cancerous cell lines, so this gene was given the name Virus-induced Rapidly Accelerated Fibrosarcoma (V-RAF). A year later, another transforming gene was found in the avian retrovirus MH2, named v-Mil - that turned out to be highly similar to v-Raf. Researchers were able to demonstrate that these genes encode enzymes th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAP2K1
Dual specificity mitogen-activated protein kinase kinase 1 is an enzyme that in humans is encoded by the ''MAP2K1'' gene. Function The protein encoded by this gene is a member of the dual-specificity protein kinase family that acts as a mitogen-activated protein (MAP) kinase kinase. MAP kinases, also known as extracellular signal-regulated kinases (ERKs), act as an integration point for multiple biochemical signals. This protein kinase lies upstream of MAP kinases and stimulates the enzymatic activity of MAP kinases upon activation by a wide variety of extra- and intracellular signals. As an essential component of the MAP kinase signal transduction pathway, this kinase is involved in many cellular processes such as proliferation, differentiation, transcription regulation and development. MAP2K1 is altered in 1.05% of all human cancers. Meiosis The genomes of diploid organisms in natural populations are highly polymorphic for insertions and deletions. During meiosis do ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
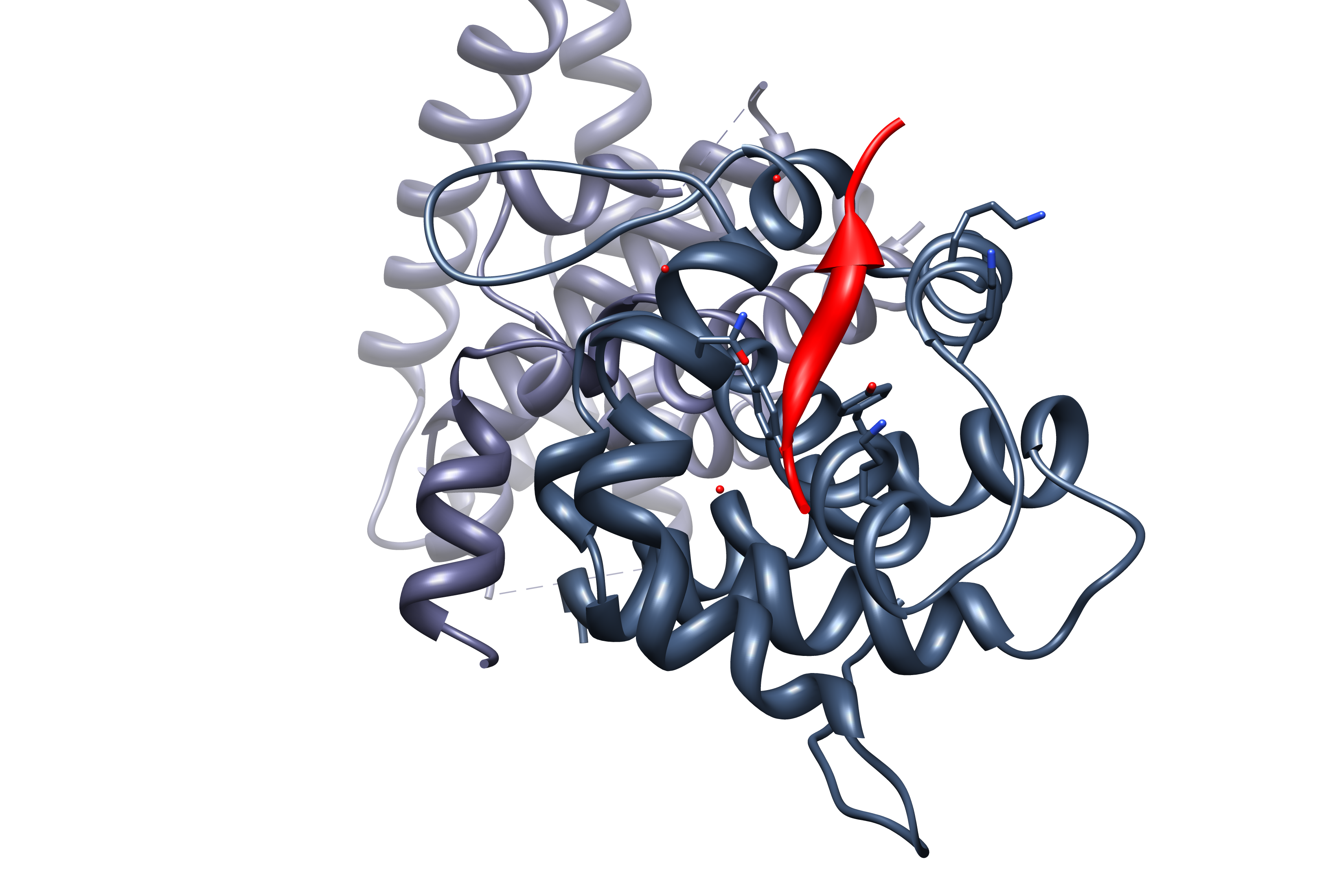
Protein Kinase Domain
The protein kinase domain is a structurally conserved protein domain containing the catalytic function of protein kinases. Protein kinases are a group of enzymes that move a phosphate group onto proteins, in a process called phosphorylation. This functions as an on/off switch for many cellular processes, including metabolism, transcription, cell cycle progression, cytoskeletal rearrangement and cell movement, apoptosis, and differentiation. They also function in embryonic development, physiological responses, and in the nervous and immune system. Abnormal phosphorylation causes many human diseases, including cancer, and drugs that affect phosphorylation can treat those diseases. Protein kinases possess a catalytic subunit which transfers the gamma phosphate from nucleoside triphosphates (almost always ATP) to the side chain of an amino acid in a protein, resulting in a conformational and/or dynamic changes affecting protein function. These enzymes fall into two broad classes, cha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
14-3-3 Protein
14-3-3 proteins are a family of conserved regulatory molecules that are expressed in all eukaryotic cells. 14-3-3 proteins have the ability to bind a multitude of functionally diverse signaling proteins, including kinases, phosphatases, and transmembrane receptors. More than 200 signaling proteins have been reported as 14-3-3 ligands. Elevated amounts of 14-3-3 proteins in cerebrospinal fluid may be a sign of Creutzfeldt–Jakob disease. Properties Seven genes encode seven distinct 14-3-3 proteins in most mammals (See ''Human genes'' below) and 13-15 genes in many higher plants, though typically in fungi they are present only in pairs. Protists have at least one. Eukaryotes can tolerate the loss of a single 14-3-3 gene if multiple genes are expressed, but deletion of all 14-3-3s (as experimentally determined in yeast) results in death. 14-3-3 proteins are structurally similar to the Tetratrico Peptide Repeat (TPR) superfamily, which generally have 9 or 10 alpha helices, and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Short Linear Motif
In molecular biology short linear motifs (SLiMs), linear motifs or minimotifs are short stretches of protein sequence that mediate protein–protein interaction. The first definition was given by Tim Hunt: "The sequences of many proteins contain short, conserved motifs that are involved in recognition and targeting activities, often separate from other functional properties of the molecule in which they occur. These motifs are linear, in the sense that three-dimensional organization is not required to bring distant segments of the molecule together to make the recognizable unit. The conservation of these motifs varies: some are highly conserved while others, for example, allow substitutions that retain only a certain pattern of charge across the motif." Attributes SLiMs are generally situated in intrinsically disordered regions (over 80% of known SLiMs), however, upon interaction with a structured partner secondary structure is often induced. The majority of annotated SLiMs con ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence (biology)
A sequence in biology is the one-dimensional ordering of monomers, covalently linked within a biopolymer; it is also referred to as the primary structure of a biological macromolecule. While it can refer to many different molecules, the term sequence is most often used to refer to a DNA sequence. See also * Protein sequence * DNA sequence * Genotype * Self-incompatibility in plants * List of geneticists * Human Genome Project * Dot plot (bioinformatics) * Multiplex Ligation-dependent Probe Amplification * Sequence analysis In bioinformatics, sequence analysis is the process of subjecting a DNA, RNA or peptide sequence to any of a wide range of analytical methods to understand its features, function, structure, or evolution. Methodologies used include sequence alig ... Molecular biology {{molecular-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L-stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure was estab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Kinase C
In cell biology, Protein kinase C, commonly abbreviated to PKC (EC 2.7.11.13), is a family of protein kinase enzymes that are involved in controlling the function of other proteins through the phosphorylation of hydroxyl groups of serine and threonine amino acid residues on these proteins, or a member of this family. PKC enzymes in turn are activated by signals such as increases in the concentration of diacylglycerol (DAG) or calcium ions (Ca2+). Hence PKC enzymes play important roles in several signal transduction cascades. In biochemistry, the PKC family consists of fifteen isozymes in humans. They are divided into three subfamilies, based on their second messenger requirements: conventional (or classical), novel, and atypical. Conventional (c)PKCs contain the isoforms α, βI, βII, and γ. These require Ca2+, DAG, and a phospholipid such as phosphatidylserine for activation. Novel (n)PKCs include the δ, ε, η, and θ isoforms, and require DAG, but do not require Ca2+ for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Zinc Finger
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (''Xenopus laevis'') transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. ''Xenopus laevis'' TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in ''Drosophila''. It often appears as a metal-binding domain in multi-domain proteins. Proteins that contain zinc fingers (zinc finger proteins) are classified into several different structural families. Unlike many other clearly defined supersecondary structures such as Greek keys or β hairpins, there are a number of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C1 Domain
C1 domain (also known as phorbol esters/diacylglycerol binding domain) binds an important secondary messenger diacylglycerol (DAG), as well as the analogous phorbol esters. Phorbol esters can directly stimulate protein kinase C, PKC. Phorbol esters (such as PMA) are analogues of DAG and potent tumor promoters that cause a variety of physiological changes when administered to both cells and tissues. DAG activates a family of serine/threonine protein kinases, collectively known as protein kinase C (PKC). Phorbol esters can directly stimulate PKC. The N-terminal region of PKC, known as C1, binds PMA and DAG in a phospholipid and zinc-dependent fashion. The C1 region contains one or two copies of a cysteine-rich domain, which is about 50 amino-acid residues long, and which is essential for DAG/PMA-binding. The DAG/PMA-binding domain binds two zinc ions; the ligands of these metal ions are probably the six cysteines and two histidines that are conserved in this domain. Human protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Raf-like Ras-binding Domain
Raf-like Ras-binding domain is an evolutionary conserved protein domain. This is the Ras-binding domain found in proteins related to Ras. Examples Human proteins containing this domain include: * ARAF * BRAF * RAF1 * RGS12, RGS14 Regulator of G-protein signaling 14 (RGS14) is a protein that in humans is encoded by the ''RGS14'' gene. Function RGS14 is a member of the regulator of G protein signalling family. This protein contains one RGS domain, two Raf-like Ras-bindi ... * TIAM1 References Protein domains {{protein-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |