|
Q-FISH Image From A Human Fibrosarcoma Cell
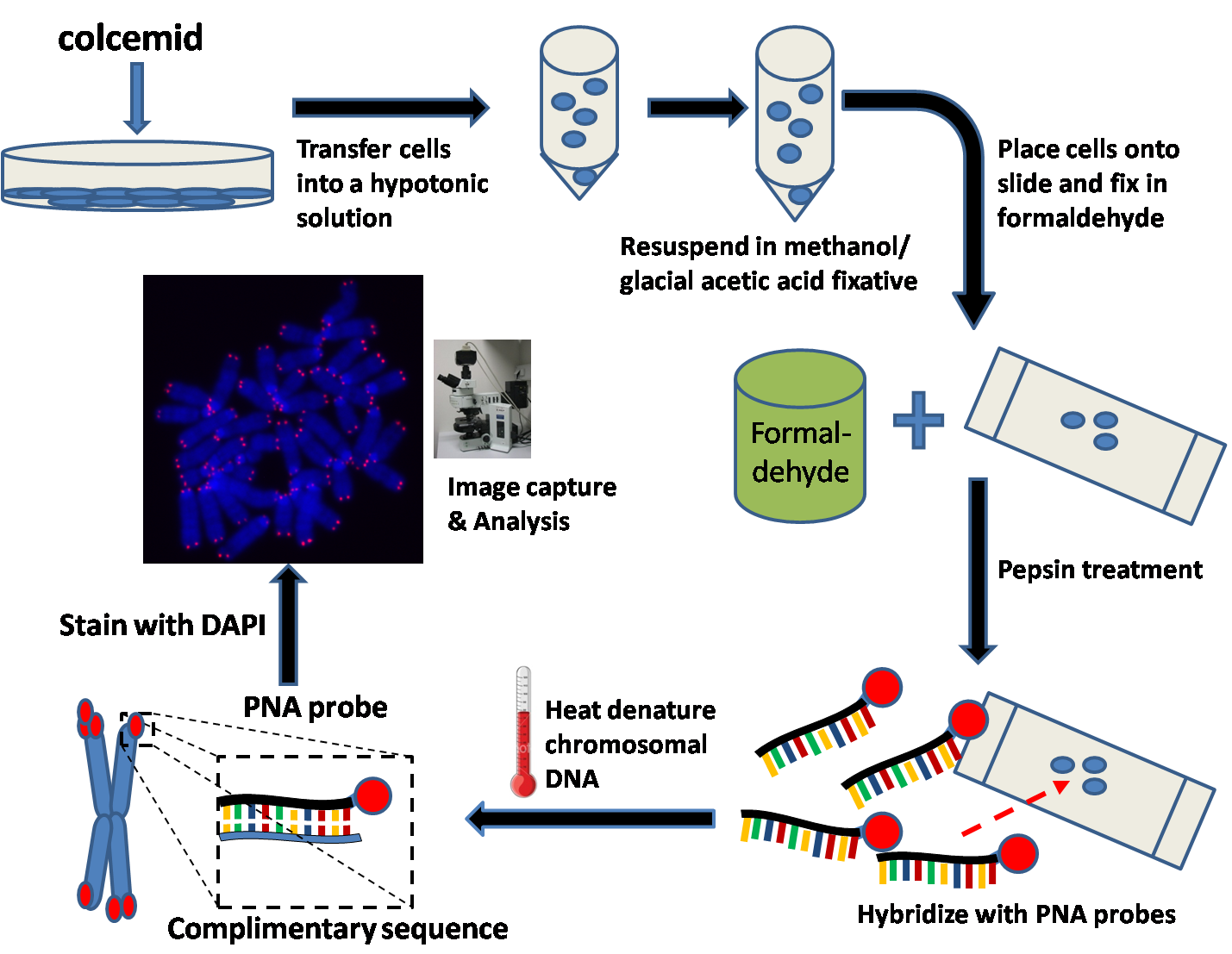
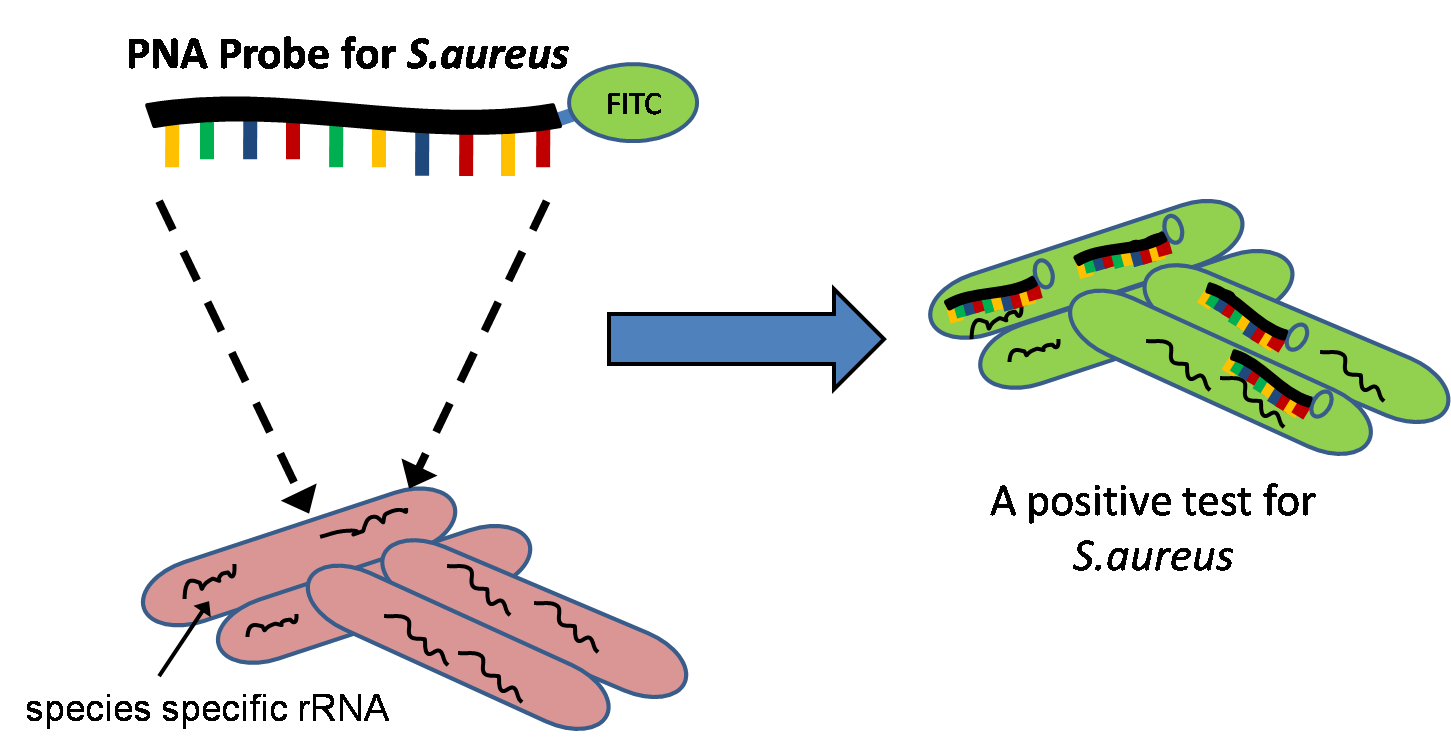
Quantitative Fluorescent ''in situ'' hybridization (Q-FISH) is a cytogenetic technique based on the traditional FISH methodology. In Q-FISH, the technique uses labelled ( Cy3 or FITC) synthetic DNA mimics called peptide nucleic acid (PNA) oligonucleotides to quantify target sequences in chromosomal DNA using fluorescent microscopy and analysis software. Q-FISH is most commonly used to study telomere length, which in vertebrates are repetitive hexameric sequences (TTAGGG) located at the distal end of chromosomes. Telomeres are necessary at chromosome ends to prevent DNA-damage responses as well as genome instability. To this day, the Q-FISH method continues to be utilized in the field of telomere research. PNAs and FISH Due to the fact that PNA backbones contain no charged phosphate groups, binding between PNA and DNA is stronger than that of DNA/DNA or DNA/RNA duplexes. Q-FISH utilizes this unique characteristic of PNAs where at low ionic strengths, PNAs can anneal to complementa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Q-FISH Image From A Human Fibrosarcoma Cell
Quantitative Fluorescent ''in situ'' hybridization (Q-FISH) is a cytogenetic technique based on the traditional FISH methodology. In Q-FISH, the technique uses labelled ( Cy3 or FITC) synthetic DNA mimics called peptide nucleic acid (PNA) oligonucleotides to quantify target sequences in chromosomal DNA using fluorescent microscopy and analysis software. Q-FISH is most commonly used to study telomere length, which in vertebrates are repetitive hexameric sequences (TTAGGG) located at the distal end of chromosomes. Telomeres are necessary at chromosome ends to prevent DNA-damage responses as well as genome instability. To this day, the Q-FISH method continues to be utilized in the field of telomere research. PNAs and FISH Due to the fact that PNA backbones contain no charged phosphate groups, binding between PNA and DNA is stronger than that of DNA/DNA or DNA/RNA duplexes. Q-FISH utilizes this unique characteristic of PNAs where at low ionic strengths, PNAs can anneal to complementa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitosis
In cell biology, mitosis () is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis gives rise to genetically identical cells in which the total number of chromosomes is maintained. Therefore, mitosis is also known as equational division. In general, mitosis is preceded by S phase of interphase (during which DNA replication occurs) and is often followed by telophase and cytokinesis; which divides the cytoplasm, organelles and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis altogether define the mitotic (M) phase of an animal cell cycle—the division of the mother cell into two daughter cells genetically identical to each other. The process of mitosis is divided into stages corresponding to the completion of one set of activities and the start of the next. These stages are preprophase (specific to plant cells), prophase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmids
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that benefit the survival of the organism and confer selective advantage such as antibiotic resistance. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain only additional genes that may be useful in certain situations or conditions. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the intern ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Locus (genetics)
In genetics, a locus (plural loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000. Genes may possess multiple variants known as alleles, and an allele may also be said to reside at a particular locus. Diploid and polyploid cells whose chromosomes have the same allele at a given locus are called homozygous with respect to that locus, while those that have different alleles at a given locus are called heterozygous. The ordered list of loci known for a particular genome is called a gene map. Gene mapping is the process of determining the specific locus or loci responsible for producing a particular phenotype or biological trait. Association mapping, also known as "linkage disequilibrium mapping", is a method of ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
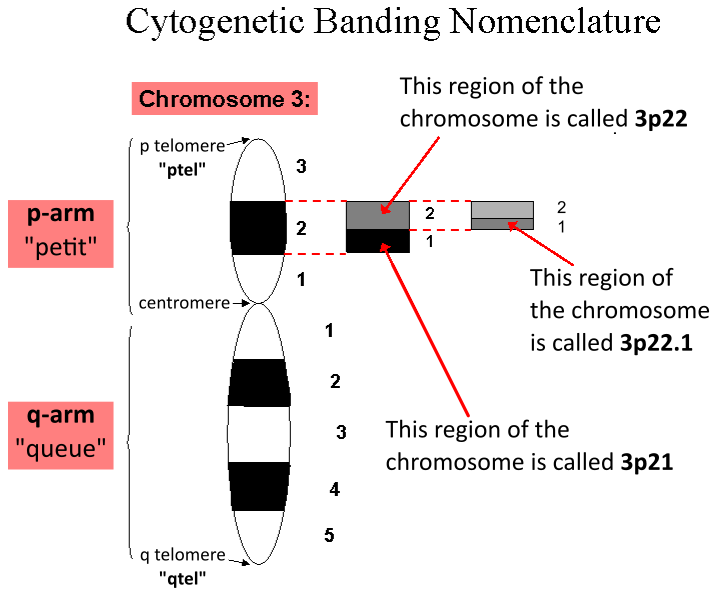
Karyotype
A karyotype is the general appearance of the complete set of metaphase chromosomes in the cells of a species or in an individual organism, mainly including their sizes, numbers, and shapes. Karyotyping is the process by which a karyotype is discerned by determining the chromosome complement of an individual, including the number of chromosomes and any abnormalities. A karyogram or idiogram is a graphical depiction of a karyotype, wherein chromosomes are organized in pairs, ordered by size and position of centromere for chromosomes of the same size. Karyotyping generally combines light microscopy and photography, and results in a photomicrographic (or simply micrographic) karyogram. In contrast, a schematic karyogram is a designed graphic representation of a karyotype. In schematic karyograms, just one of the sister chromatids of each chromosome is generally shown for brevity, and in reality they are generally so close together that they look as one on photomicrographs as well ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Photobleaching
In optics, photobleaching (sometimes termed fading) is the photochemical alteration of a dye or a fluorophore molecule such that it is permanently unable to fluoresce. This is caused by cleaving of covalent bonds or non-specific reactions between the fluorophore and surrounding molecules. Such irreversible modifications in covalent bonds are caused by transition from a singlet state to the triplet state of the fluorophores. The number of excitation cycles to achieve full bleaching varies. In microscopy, photobleaching may complicate the observation of fluorescent molecules, since they will eventually be destroyed by the light exposure necessary to stimulate them into fluorescing. This is especially problematic in time-lapse microscopy. However, photobleaching may also be used prior to applying the (primarily antibody-linked) fluorescent molecules, in an attempt to quench autofluorescence. This can help improve the signal-to-noise ratio. Photobleaching may also be exploited to study ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DAPI
DAPI (pronounced 'DAPPY', /ˈdæpiː/), or 4′,6-diamidino-2-phenylindole, is a fluorescent stain that binds strongly to adenine–thymine-rich regions in DNA. It is used extensively in fluorescence microscopy. As DAPI can pass through an intact cell membrane, it can be used to stain both live and fixed cells, though it passes through the membrane less efficiently in live cells and therefore provides a marker for membrane viability. History DAPI was first synthesised in 1971 in the laboratory of Otto Dann as part of a search for drugs to treat trypanosomiasis. Although it was unsuccessful as a drug, further investigation indicated it bound strongly to DNA and became more fluorescent when bound. This led to its use in identifying mitochondrial DNA in ultracentrifugation in 1975, the first recorded use of DAPI as a fluorescent DNA stain. Strong fluorescence when bound to DNA led to the rapid adoption of DAPI for fluorescent staining of DNA for fluorescence microscopy. Its use f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Denaturation (biochemistry)
In biochemistry, denaturation is a process in which proteins or nucleic acids lose the quaternary structure, tertiary structure, and secondary structure which is present in their native state, by application of some external stress or compound such as a strong acid or base, a concentrated inorganic salt, an organic solvent (e.g., alcohol or chloroform), agitation and radiation or heat. If proteins in a living cell are denatured, this results in disruption of cell activity and possibly cell death. Protein denaturation is also a consequence of cell death. Denatured proteins can exhibit a wide range of characteristics, from conformational change and loss of solubility to aggregation due to the exposure of hydrophobic groups. The loss of solubility as a result of denaturation is called ''coagulation''. Denatured proteins lose their 3D structure and therefore cannot function. Protein folding is key to whether a globular or membrane protein can do its job correctly; it must be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Hierarchy of proteases Based on catalytic residue Proteases can be classified into seven broad groups: * Serine protease ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pepsin
Pepsin is an endopeptidase that breaks down proteins into smaller peptides. It is produced in the gastric chief cells of the stomach lining and is one of the main digestive enzymes in the digestive systems of humans and many other animals, where it helps digest the proteins in food. Pepsin is an aspartic protease, using a catalytic aspartate in its active site. It is one of three principal endopeptidases (enzymes cutting proteins in the middle) in the human digestive system, the other two being chymotrypsin and trypsin. There are also exopeptidases which remove individual amino acids at both ends of proteins (carboxypeptidases produced by the pancreas and aminopeptidases secreted by the small intestine). During the process of digestion, these enzymes, each of which is specialized in severing links between particular types of amino acids, collaborate to break down dietary proteins into their components, i.e., peptides and amino acids, which can be readily absorbed by the sma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fixation (histology)
In the fields of histology, pathology, and cell biology, fixation is the preservation of biological tissues from decay due to autolysis or putrefaction. It terminates any ongoing biochemical reactions and may also increase the treated tissues' mechanical strength or stability. Tissue fixation is a critical step in the preparation of histological sections, its broad objective being to preserve cells and tissue components and to do this in such a way as to allow for the preparation of thin, stained sections. This allows the investigation of the tissues' structure, which is determined by the shapes and sizes of such macromolecules (in and around cells) as proteins and nucleic acids. Purposes In performing their protective role, fixatives denature proteins by coagulation, by forming additive compounds, or by a combination of coagulation and additive processes. A compound that adds chemically to macromolecules stabilizes structure most effectively if it is able to combine with part ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Formaldehyde
Formaldehyde ( , ) (systematic name methanal) is a naturally occurring organic compound with the formula and structure . The pure compound is a pungent, colourless gas that polymerises spontaneously into paraformaldehyde (refer to section Forms below), hence it is stored as an aqueous solution (formalin), which is also used to store animal specimens. It is the simplest of the aldehydes (). The common name of this substance comes from its similarity and relation to formic acid. Formaldehyde is an important precursor to many other materials and chemical compounds. In 1996, the installed capacity for the production of formaldehyde was estimated at 8.7 million tons per year. It is mainly used in the production of industrial resins, e.g., for particle board and coatings. Forms Formaldehyde is more complicated than many simple carbon compounds in that it adopts several diverse forms. These compounds can often be used interchangeably and can be interconverted. *Molecular formald ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |