|
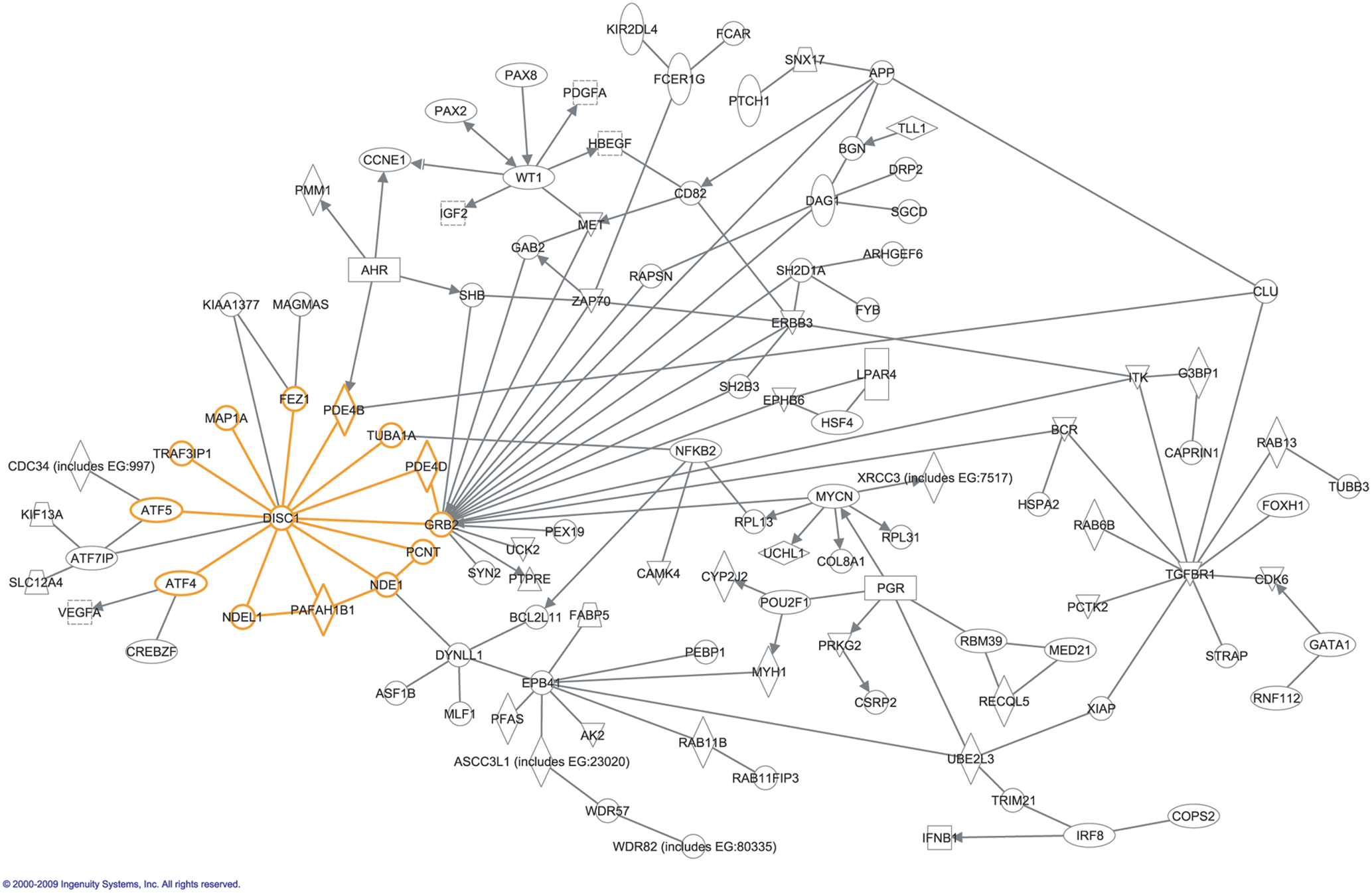
Protein–protein Interaction Prediction
Protein–protein interaction prediction is a field combining bioinformatics and structural biology in an attempt to identify and catalog physical interactions between pairs or groups of proteins. Understanding protein–protein interactions is important for the investigation of intracellular signaling pathways, modelling of protein complex structures and for gaining insights into various biochemical processes. ''Experimentally'', physical interactions between pairs of proteins can be inferred from a variety of techniques, including yeast two-hybrid systems, protein-fragment complementation assays (PCA), affinity purification/mass spectrometry, protein microarrays, fluorescence resonance energy transfer (FRET), and Microscale Thermophoresis (MST). Efforts to experimentally determine the interactome of numerous species are ongoing. Experimentally determined interactions usually provide the basis for ''computational methods'' to predict interactions, e.g. using homologous protein se ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for '' in silico'' analyses of biological queries using computational and statistical techniques. Bioinformatics includes biological studies that use computer programming as part of their methodology, as well as specific analysis "pipelines" that are repeatedly used, particularly in the field of genomics. Common uses of bioinformatics include the identification of candidates genes and single nucleotide polymorphisms (SNPs). Often, such identification is made with the aim to better understand the genetic basis of disease, unique adaptations, desirable properties (e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SH3 Domain
The SRC Homology 3 Domain (or SH3 domain) is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src. It has also been identified in several other protein families such as: PI3 Kinase, Ras GTPase-activating protein, CDC24 and cdc25. SH3 domains are found in proteins of signaling pathways regulating the cytoskeleton, the Ras protein, and the Src kinase and many others. The SH3 proteins interact with adaptor proteins and tyrosine kinases. Interacting with tyrosine kinases, SH3 proteins usually bind far away from the active site. Approximately 300 SH3 domains are found in proteins encoded in the human genome. In addition to that, the SH3 domain was responsible for controlling protein-protein interactions in the signal transduction pathways and regulating the interactions ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure Prediction
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its secondary and tertiary structure from primary structure. Structure prediction is different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by computational biology; and it is important in medicine (for example, in drug design) and biotechnology (for example, in the design of novel enzymes). Starting in 1994, the performance of current methods is assessed biannually in the CASP experiment (Critical Assessment of Techniques for Protein Structure Prediction). A continuous evaluation of protein structure prediction web servers is performed by the community project CAMEO3D. Protein structure and terminology Proteins are chains of amino acids joined together by peptide bonds. Many conformations of this chain are possible due to the rotation of the main chain abou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Function Prediction
Protein function prediction methods are techniques that bioinformatics researchers use to assign biological or biochemical roles to proteins. These proteins are usually ones that are poorly studied or predicted based on genomic sequence data. These predictions are often driven by data-intensive computational procedures. Information may come from nucleic acid sequence homology, gene expression profiles, protein domain structures, text mining of publications, phylogenetic profiles, phenotypic profiles, and protein-protein interaction. Protein function is a broad term: the roles of proteins range from catalysis of biochemical reactions to transport to signal transduction, and a single protein may play a role in multiple processes or cellular pathways. Generally, function can be thought of as, "anything that happens to or through a protein". The Gene Ontology Consortium provides a useful classification of functions, based on a dictionary of well-defined terms divided into three mai ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein–protein Interaction
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and the hydrophobic effect. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context. Proteins rarely act alone as their functions tend to be regulated. Many molecular processes within a cell are carried out by molecular machines that are built from numerous protein components organized by their PPIs. These physiological interactions make up the so-called interactomics of the organism, while aberrant PPIs are the basis of multiple aggregation-related diseases, such as Creutzfeldt–Jakob and Alzheimer's diseases. PPIs have been studied with many methods and from different perspectives: biochemistry, quantum chemistry, molecular dynamics, signal trans ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Interactome
In molecular biology, an interactome is the whole set of molecular interactions in a particular cell. The term specifically refers to physical interactions among molecules (such as those among proteins, also known as protein–protein interactions, PPIs; or between small molecules and proteins) but can also describe sets of indirect interactions among genes (genetic interactions). The word "interactome" was originally coined in 1999 by a group of French scientists headed by Bernard Jacq. Mathematically, interactomes are generally displayed as graphs. Though interactomes may be described as biological networks, they should not be confused with other networks such as neural networks or food webs. Molecular interaction networks Molecular interactions can occur between molecules belonging to different biochemical families (proteins, nucleic acids, lipids, carbohydrates, etc.) and also within a given family. Whenever such molecules are connected by physical interactions, they form molecu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein-protein Docking
Macromolecular docking is the computational modelling of the quaternary structure of complexes formed by two or more interacting biological macromolecules. Protein–protein complexes are the most commonly attempted targets of such modelling, followed by protein–nucleic acid complexes. The ultimate goal of docking is the prediction of the three-dimensional structure of the macromolecular complex of interest as it would occur in a living organism. Docking itself only produces plausible candidate structures. These candidates must be ranked using methods such as scoring functions to identify structures that are most likely to occur in nature. The term "docking" originated in the late 1970s, with a more restricted meaning; then, "docking" meant refining a model of a complex structure by optimizing the separation between the interactors but keeping their relative orientations fixed. Later, the relative orientations of the interacting partners in the modelling was allowed to vary, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bayesian Method
Bayesian inference is a method of statistical inference in which Bayes' theorem is used to update the probability for a hypothesis as more evidence or information becomes available. Bayesian inference is an important technique in statistics, and especially in mathematical statistics. Bayesian updating is particularly important in the dynamic analysis of a sequence of data. Bayesian inference has found application in a wide range of activities, including science, engineering, philosophy, medicine, sport, and law. In the philosophy of decision theory, Bayesian inference is closely related to subjective probability, often called "Bayesian probability". Introduction to Bayes' rule Formal explanation Bayesian inference derives the posterior probability as a consequence of two antecedents: a prior probability and a "likelihood function" derived from a statistical model for the observed data. Bayesian inference computes the posterior probability according to Bayes' theorem: P(H\ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Van Der Waals Radius
The van der Waals radius, ''r'', of an atom is the radius of an imaginary hard sphere representing the distance of closest approach for another atom. It is named after Johannes Diderik van der Waals, winner of the 1910 Nobel Prize in Physics, as he was the first to recognise that atoms were not simply points and to demonstrate the physical consequences of their size through the van der Waals equation of state. van der Waals volume The van der Waals volume, ''V'', also called the atomic volume or molecular volume, is the atomic property most directly related to the van der Waals radius. It is the volume "occupied" by an individual atom (or molecule). The van der Waals volume may be calculated if the van der Waals radii (and, for molecules, the inter-atomic distances, and angles) are known. For a single atom, it is the volume of a sphere whose radius is the van der Waals radius of the atom: V_ = \pi r_^3. For a molecule, it is the volume enclosed by the van der Waals surfac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations (PDBe, PDBj, RCSB, and BMRB). The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB. The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other databases use protein structures deposited in the PDB. For example, SCOP and CATH classify protein structures, while PDBsum provides a graphic overview of PDB entries using information from other sources, such as Gene ontology. History Two force ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Interolog
An interolog is a conserved interaction between a pair of proteins which have interacting Homology (biology), homologs in another organism. The term was introduced in a 2000 paper by Walhout et al. Example Suppose that A and B are two different interacting human proteins, and A' and B' are two different interacting dog proteins. Then the interaction between A and B is an interolog of the interaction between A' and B' if the following conditions all hold: *A is a homolog of A'. (Protein homologs have similar amino acid sequences and derive from a common ancestral sequence). *B is a homolog of B'. *A and B interact. *A' and B' interact. Thus, interologs are homologous pairs of protein interactions across different organisms. See also *Homology (biology) *Systems biology *Bioinformatics References * * * External links Interactome.org Interactome portal site. Interactomics.org Interactomics portal site. Cross-species interaction prediction site. Protein complexes Bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tryptophan Synthase
Tryptophan synthase or tryptophan synthetase is an enzyme () that catalyses the final two steps in the biosynthesis of tryptophan. It is commonly found in Eubacteria, Archaebacteria, Protista, Fungi, and Plantae. However, it is absent from Animalia. It is typically found as an α2β2 tetramer. The α subunits catalyze the reversible formation of indole and glyceraldehyde-3-phosphate (G3P) from indole-3-glycerol phosphate (IGP). The β subunits catalyze the irreversible condensation of indole and serine to form tryptophan in a pyridoxal phosphate (PLP) dependent reaction. Each α active site is connected to a β active site by a 25 angstrom long hydrophobic channel contained within the enzyme. This facilitates the diffusion of indole formed at α active sites directly to β active sites in a process known as substrate channeling. The active sites of tryptophan synthase are allosterically coupled. Enzyme structure Subunits: Tryptophan synthase typically exists as an α-ββ-α comple ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |