Protein function prediction on:
[Wikipedia]
[Google]
[Amazon]
Protein function prediction methods are techniques that
 Proteins of similar sequence are usually homologous and thus have a similar function. Hence proteins in a newly sequenced
Proteins of similar sequence are usually homologous and thus have a similar function. Hence proteins in a newly sequenced
 Because 3D
Because 3D

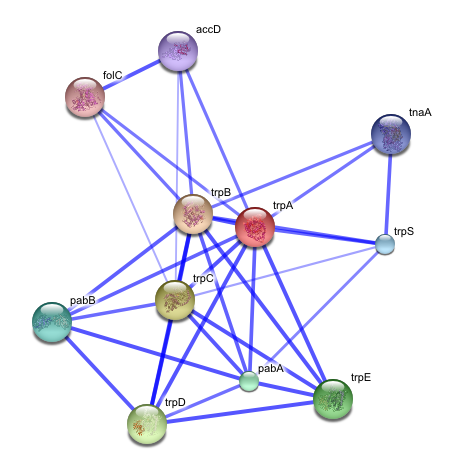 Guilt by association type algorithms may be used to produce a functional association network for a given target group of genes or proteins. These networks serve as a representation of the evidence for shared/similar function within a group of genes, where nodes represent genes/proteins and are linked to each other by edges representing evidence of shared function.
Guilt by association type algorithms may be used to produce a functional association network for a given target group of genes or proteins. These networks serve as a representation of the evidence for shared/similar function within a group of genes, where nodes represent genes/proteins and are linked to each other by edges representing evidence of shared function.
VisANT
Visual analysis of networks and integrative visual data-mining.
Mantis
A consensus-driven function prediction tool that dynamically integrates multiple reference databases.
The dcGO database
Protein Data Bank
Catalytic Site Atlas
RaptorX Server for model-assisted protein function prediction
* Blast2GO, high-throughput tool for protein function prediction and functional annotation
webpage
. {{DEFAULTSORT:Protein Function Prediction Bioinformatics Protein methods
bioinformatics
Bioinformatics () is an interdisciplinary field of science that develops methods and Bioinformatics software, software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, ...
researchers use to assign biological or biochemical roles to protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s. These proteins are usually ones that are poorly studied or predicted based on genomic sequence data. These predictions are often driven by data-intensive computational procedures. Information may come from nucleic acid sequence homology
Sequence homology is the homology (biology), biological homology between DNA sequence, DNA, RNA sequence, RNA, or Protein primary structure, protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments ...
, gene expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, ...
profiles, protein domain
In molecular biology, a protein domain is a region of a protein's Peptide, polypeptide chain that is self-stabilizing and that Protein folding, folds independently from the rest. Each domain forms a compact folded Protein tertiary structure, thre ...
structures, text mining
Text mining, text data mining (TDM) or text analytics is the process of deriving high-quality information from text. It involves "the discovery by computer of new, previously unknown information, by automatically extracting information from differe ...
of publications, phylogenetic profiles, phenotypic profiles, and protein-protein interaction. Protein function is a broad term: the roles of proteins range from catalysis of biochemical reactions to transport to signal transduction
Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a biochemical cascade, series of molecular events. Proteins responsible for detecting stimuli are generally termed receptor (biology), rece ...
, and a single protein may play a role in multiple processes or cellular pathways.
Generally, function can be thought of as, "anything that happens to or through a protein". The Gene Ontology Consortium provides a useful classification of functions, based on a dictionary of well-defined terms divided into three main categories of ''molecular function, biological process'' and ''cellular component''. Researchers can query this database with a protein name or accession number to retrieve associated Gene Ontology (GO) terms or annotations based on computational or experimental evidence.
While techniques such as microarray
A microarray is a multiplex (assay), multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a Substrate (materials science), solid substrate—usu ...
analysis, RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by ...
, and the yeast two-hybrid system can be used to experimentally demonstrate the function of a protein, advances in sequencing technologies have made the rate at which proteins can be experimentally characterized much slower than the rate at which new sequences become available. Thus, the annotation of new sequences is mostly by ''prediction'' through computational methods, as these types of annotation can often be done quickly and for many genes or proteins at once. The first such methods inferred function based on homologous proteins with known functions (homology-based function prediction). The development of context-based and structure based methods have expanded what information can be predicted, and a combination of methods can now be used to get a picture of complete cellular pathways based on sequence data. The importance and prevalence of computational prediction of gene function is underlined by an analysis of 'evidence codes' used by the GO database: as of 2010, 98% of annotations were listed under the code IEA (inferred from electronic annotation) while only 0.6% were based on experimental evidence.
Homology-based methods
genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
are routinely annotated using the sequences of similar proteins in related genomes.
However, closely related proteins do not always share the same function. For example, the yeast Gal1 and Gal3 proteins are paralog
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a sp ...
s (73% identity and 92% similarity) that have evolved very different functions with Gal1 being a galactokinase and Gal3 being a transcriptional inducer.
There is no hard sequence-similarity threshold for "safe" function prediction; many proteins of barely detectable sequence similarity have the same function while others (such as Gal1 and Gal3) are highly similar but have evolved different functions. As a rule of thumb, sequences that are more than 30-40% identical are usually considered as having the same or a very similar function.
For enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
s, predictions of specific functions are especially difficult, as they only need a few key residues in their active site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the ''binding s ...
, hence very different sequences can have very similar activities. By contrast, even with sequence identity of 70% or greater, 10% of any pair of enzymes have different substrates; and differences in the actual enzymatic reactions are not uncommon near 50% sequence identity.
Sequence motif-based methods
The development of protein domain databases such asPfam
Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models. The latest version of Pfam, 37.0, was released in June 2024 and contains 21,979 families. It is cur ...
(Protein Families Database) allow us to find known domains within a query sequence, providing evidence for likely functions. The dcGO website contains annotations to both the individual domains and supra-domains (i.e., combinations of two or more successive domains), thus via dcGO Predictor allowing for the function predictions in a more realistic manner. Within protein domain
In molecular biology, a protein domain is a region of a protein's Peptide, polypeptide chain that is self-stabilizing and that Protein folding, folds independently from the rest. Each domain forms a compact folded Protein tertiary structure, thre ...
s, shorter signatures known as ''motifs'' are associated with particular functions, and motif databases such as PROSITE ('database of protein domains, families and functional sites') can be searched using a query sequence. Motifs can, for example, be used to predict subcellular localization The cells of eukaryotic organisms are elaborately subdivided into functionally-distinct membrane-bound compartments. Some major constituents of eukaryotic cells are: extracellular space, plasma membrane, cytoplasm, nucleus, mitochondria, Golgi a ...
of a protein (where in the cell the protein is sent after synthesis). Short signal peptides direct certain proteins to a particular location such as the mitochondria, and various tools exist for the prediction of these signals in a protein sequence. For example, SignalP, which has been updated several times as methods are improved.
Thus, aspects of a protein's function can be predicted without comparison to other full-length homologous protein sequences.
Structure-based methods
 Because 3D
Because 3D protein structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, which are the monomers of the polymer. A single amino acid ...
is generally more well conserved than protein sequence, structural similarity is a good indicator of similar function in two or more proteins. Many programs have been developed to screen a known protein structure against the Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules such as proteins and nucleic acids, which is overseen by the Worldwide Protein Data Bank (wwPDB). This structural data is obtained a ...
and report similar structures (for example, FATCAT (Flexible structure AlignmenT by Chaining AFPs (Aligned Fragment Pairs) with Twists), CE (combinatorial extension)) and DeepAlign (protein structure alignment beyond spatial proximity). Similarly, the main protein databases, such as UniProt
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived fro ...
, have built-in tools to search any given protein sequences against structure databases, and link to related proteins of known structure.
Protein structure prediction
To deal with the situation that many protein sequences have no solved structures, some function prediction servers such as RaptorX are also developed that can first predict the 3D model of a sequence and then use structure-based method to predict functions based upon the predicted 3D model. In many cases instead of the whole protein structure, the 3D structure of a particular motif representing anactive site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the ''binding s ...
or binding site can be targeted. The Structurally Aligned Local Sites of Activity (SALSA) method, developed by Mary Jo Ondrechen and students, utilizes computed chemical properties of the individual amino acids to identify local biochemically active sites. Databases such as Catalytic Site Atlas have been developed that can be searched using novel protein sequences to predict specific functional sites.
Computational solvent mapping
One of the challenges involved in protein function prediction is discovery of the active site. This is complicated by certain active sites not being formed – essentially existing – until the protein undergoes conformational changes brought on by the binding of small molecules. Most protein structures have been determined byX-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
which requires a purified protein crystal
A crystal or crystalline solid is a solid material whose constituents (such as atoms, molecules, or ions) are arranged in a highly ordered microscopic structure, forming a crystal lattice that extends in all directions. In addition, macros ...
. As a result, existing structural models are generally of a purified protein and as such lack the conformational changes that are created when the protein interacts with small molecules.
Computational solvent mapping utilizes probes (small organic molecules) that are computationally 'moved' over the surface of the protein searching for sites where they tend to cluster. Multiple different probes are generally applied with the goal being to obtain a large number of different protein-probe conformations. The generated clusters are then ranked based on the cluster's average free energy. After computationally mapping multiple probes, the site of the protein where relatively large numbers of clusters form typically corresponds to an active site on the protein.
This technique is a computational adaptation of 'wet lab' work from 1996. It was discovered that ascertaining the structure of a protein while it is suspended in different solvents and then superimposing those structures on one another produces data where the organic solvent molecules (that the proteins were suspended in) typically cluster at the protein's active site. This work was carried out as a response to realizing that water molecules are visible in the electron density maps produced by X-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
. The water molecules are interacting with the protein and tend to cluster at the protein's polar regions. This led to the idea of immersing the purified protein crystal in other solvents (e.g. ethanol
Ethanol (also called ethyl alcohol, grain alcohol, drinking alcohol, or simply alcohol) is an organic compound with the chemical formula . It is an Alcohol (chemistry), alcohol, with its formula also written as , or EtOH, where Et is the ps ...
, isopropanol
Isopropyl alcohol (IUPAC name propan-2-ol and also called isopropanol or 2-propanol) is a colorless, flammable, organic compound with a pungent alcoholic odor.
Isopropyl alcohol, an organic polar molecule, is miscible in water, ethanol, an ...
, etc.) to determine where these molecules cluster on the protein. The solvents can be chosen based on what they approximate, that is, what molecule this protein may interact with (e.g. ethanol
Ethanol (also called ethyl alcohol, grain alcohol, drinking alcohol, or simply alcohol) is an organic compound with the chemical formula . It is an Alcohol (chemistry), alcohol, with its formula also written as , or EtOH, where Et is the ps ...
can probe for interactions with the amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
serine
Serine
(symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − ...
, isopropanol
Isopropyl alcohol (IUPAC name propan-2-ol and also called isopropanol or 2-propanol) is a colorless, flammable, organic compound with a pungent alcoholic odor.
Isopropyl alcohol, an organic polar molecule, is miscible in water, ethanol, an ...
a probe for threonine
Threonine (symbol Thr or T) is an amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form when dissolved in water), a carboxyl group (which is in the deprotonated −COO− ...
, etc.). It is vital that the protein crystal maintains its tertiary structure
Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the ...
in each solvent. This process is repeated for multiple solvents and then this data can be used to try to determine potential active sites on the protein. Ten years later this technique was developed into an algorithm by Clodfelter et al.
Genome context-based methods
Many of the newer methods for protein function prediction are not based on comparison of sequence or structure as above, but on some type of correlation between novel genes/proteins and those that already have annotations. Several methods have been developed to predict gene function on the local genomic orphylogenomic
Phylogenomics is the intersection of the fields of evolution and genomics. The term has been used in multiple ways to refer to analysis that involves genome data and evolutionary reconstructions. It is a group of techniques within the larger fields ...
context and structure of genes:
Phylogenetic profiling is based on the observation that two or more proteins with the same pattern of presence or absence in many different genomes most likely have a functional link. Whereas homology-based methods can often be used to identify molecular functions of a protein, context-based approaches can be used to predict cellular function, or the biological process in which a protein acts. For example, proteins involved in the same metabolic pathway are likely to be present in a genome together or are absent altogether, suggesting that these genes work together in a functional context.

Operon
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splic ...
s are clusters of genes that are transcribed together. Based on co-transcription data but also based on the fact that the order of genes in operons is often conserved across many bacteria, indicates that they act together.
Gene fusion
In genetics, a fusion gene is a hybrid gene formed from two previously independent genes. It can occur as a result of translocation, interstitial deletion, or chromosomal inversion. Fusion genes have been found to be prevalent in all main types ...
occurs when two or more genes encode two or more proteins in one organism and have, through evolution, combined to become a single gene in another organism (or vice versa for ''gene fission''). This concept has been used, for example, to search all ''E. coli
''Escherichia coli'' ( )Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escherichia'' that is commonly foun ...
'' protein sequences for homology in other genomes and find over 6000 pairs of sequences with shared homology to single proteins in another genome, indicating potential interaction between each of the pairs. Because the two sequences in each protein pair are non-homologous, these interactions could not be predicted using homology-based methods.
Gene expression and location-based methods
Inprokaryote
A prokaryote (; less commonly spelled procaryote) is a unicellular organism, single-celled organism whose cell (biology), cell lacks a cell nucleus, nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Gree ...
s, clusters of genes that are physically close together in the genome often conserve together through evolution, and tend to encode proteins that interact or are part of the same operon
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splic ...
. Thus, ''chromosomal proximity'' also called the gene neighbour method can be used to predict functional similarity between proteins, at least in prokaryotes. Chromosomal proximity has also been seen to apply for some pathways in selected eukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
genomes, including ''Homo sapiens'', and with further development gene neighbor methods may be valuable for studying protein interactions in eukaryotes.
Genes involved in similar functions are also often co-transcribed, so that an unannotated protein can often be predicted to have a related function to proteins with which it co-expresses. The guilt by association algorithm
In mathematics and computer science, an algorithm () is a finite sequence of Rigour#Mathematics, mathematically rigorous instructions, typically used to solve a class of specific Computational problem, problems or to perform a computation. Algo ...
s developed based on this approach can be used to analyze large amounts of sequence data and identify genes with expression patterns similar to those of known genes. Often, a guilt by association study compares a group of candidate gene
The candidate gene approach to conducting genetic association studies focuses on associations between genetic variation within pre-specified genes of interest, and Phenotype (clinical medicine), phenotypes or disease states. This is in contrast to ...
s (unknown function) to a target group (for example, a group of genes known to be associated with a particular disease), and rank the candidate genes by their likelihood of belonging to the target group based on the data. Based on recent studies, however, it has been suggested that some problems exist with this type of analysis. For example, because many proteins are multifunctional, the genes encoding them may belong to several target groups. It is argued that such genes are more likely to be identified in guilt by association studies, and thus predictions are not specific.
With the accumulation of RNA-seq data that are capable of estimating expression profiles for alternatively spliced isoforms, machine learning algorithms have also been developed for predicting and differentiating functions at the isoform level. This represents an emerging research area in function prediction, which integrates large-scale, heterogeneous genomic data to infer functions at the isoform level.
Network-based methods
 Guilt by association type algorithms may be used to produce a functional association network for a given target group of genes or proteins. These networks serve as a representation of the evidence for shared/similar function within a group of genes, where nodes represent genes/proteins and are linked to each other by edges representing evidence of shared function.
Guilt by association type algorithms may be used to produce a functional association network for a given target group of genes or proteins. These networks serve as a representation of the evidence for shared/similar function within a group of genes, where nodes represent genes/proteins and are linked to each other by edges representing evidence of shared function.
Integrated networks
Several networks based on different data sources can be combined into a composite network, which can then be used by a prediction algorithm to annotate candidate genes or proteins. For example, the developers of the bioPIXIE system used a wide variety of ''Saccharomyces cerevisiae'' (yeast) genomic data to produce a composite functional network for that species. This resource allows the visualization of known networks representing biological processes, as well as the prediction of novel components of those networks. Many algorithms have been developed to predict function based on the integration of several data sources (e.g. genomic, proteomic, protein interaction, etc.), and testing on previously annotated genes indicates a high level of accuracy. Disadvantages of some function prediction algorithms have included a lack of accessibility, and the time required for analysis. Faster, more accurate algorithms such as GeneMANIA (multiple association network integration algorithm) have however been developed in recent years and are publicly available on the web, indicating the future direction of function prediction.Tools and databases for protein function prediction
STRING
String or strings may refer to:
*String (structure), a long flexible structure made from threads twisted together, which is used to tie, bind, or hang other objects
Arts, entertainment, and media Films
* ''Strings'' (1991 film), a Canadian anim ...
: web tool that integrates various data sources for function prediction.VisANT
Visual analysis of networks and integrative visual data-mining.
Mantis
A consensus-driven function prediction tool that dynamically integrates multiple reference databases.
See also
*Gene prediction
In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. This includes protein-coding genes as well as RNA genes, but may also include prediction of other functio ...
*Protein–protein interaction prediction Protein–protein interaction prediction is a field combining bioinformatics and structural biology in an attempt to identify and catalog physical interactions between pairs or groups of proteins. Understanding protein–protein interactions is impo ...
*Protein structure prediction
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its Protein secondary structure, secondary and Protein tertiary structure, tertiary structure ...
*Structural genomics
Structural genomics seeks to describe the Protein Structure, 3-dimensional structure of every protein encoded by a given genome. This genome-based approach allows for a high-throughput method of structure determination by a combination of protein ...
*Functional genomics
Functional genomics is a field of molecular biology that attempts to describe gene (and protein) functions and interactions. Functional genomics make use of the vast data generated by genomic and transcriptomic projects (such as genome sequen ...
References
External links
The dcGO database
Protein Data Bank
Catalytic Site Atlas
RaptorX Server for model-assisted protein function prediction
* Blast2GO, high-throughput tool for protein function prediction and functional annotation
webpage
. {{DEFAULTSORT:Protein Function Prediction Bioinformatics Protein methods