|
Post Transcriptional Modification
Transcriptional modification or co-transcriptional modification is a set of biological processes common to most eukaryotic cells by which an RNA primary transcript is chemically altered following transcription from a gene to produce a mature, functional RNA molecule that can then leave the nucleus and perform any of a variety of different functions in the cell. There are many types of post-transcriptional modifications achieved through a diverse class of molecular mechanisms. One example is the conversion of precursor messenger RNA transcripts into mature messenger RNA that is subsequently capable of being translated into protein. This process includes three major steps that significantly modify the chemical structure of the RNA molecule: the addition of a 5' cap, the addition of a 3' polyadenylated tail, and RNA splicing. Such processing is vital for the correct translation of eukaryotic genomes because the initial precursor mRNA produced by transcription often contains both exon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacteria and Archaea (both prokaryotes) make up the other two domains. The eukaryotes are usually now regarded as having emerged in the Archaea or as a sister of the Asgard archaea. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among archaea. Eukaryotes represent a small minority of the number of organisms, but, due to their generally much larger size, their collective global biomass is estimated to be about equal to that of prokaryotes. Eukaryotes emerged approximately 2.3–1.8 billion years ago, during the Proterozoic eon, likely as flagellated phagotrophs. Their name comes from the Greek εὖ (''eu'', "well" or "good") and κάρυον (''karyon'', "nut" or "kernel"). Euka ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
7-methylguanosine
7-Methylguanosine (m7G) is a modified purine nucleoside. It is a methylated version of guanosine and when found in human urine, it may be a biomarker of some types of cancer. In the RNAs, 7-methylguanosine have been used to study and examine the reaction evolving methylguanosine. It also plays a role in mRNA as a blocking group at its 5´-end. See also *METTL1 *23S rRNA (guanine2445-N2)-methyltransferase *16S rRNA (guanine527-N7)-methyltransferase 16S rRNA (guanine527-N7)-methyltransferase (, ''ribosomal RNA small subunit methyltransferase G'', ''16S rRNA methyltransferase RsmG'', ''GidB'', ''rsmG (gene)'') is an enzyme with systematic name ''S-adenosyl-L-methionine:16S rRNA (guanine527-N7)- ... References External links Metabocard for 7-Methylguanosine (HMDB01107) Human Metabolome Database, University of Alberta Nucleosides Purines Hydroxymethyl compounds Biomarkers {{organic-compound-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Poly(A) Tail
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature mRNA for translation. In many bacteria, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of gene expression. The process of polyadenylation begins as the transcription of a gene terminates. The 3′-most segment of the newly made pre-mRNA is first cleaved off by a set of proteins; these proteins then synthesize the poly(A) tail at the RNA's 3′ end. In some genes these proteins add a poly(A) tail at one of several possible sites. Therefore, polyadenylation can produce more than one transcript from a single gene (alternative polyadenylation), similar to alternative splicing. The poly(A) tail is important for the nucle ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenine
Adenine () ( symbol A or Ade) is a nucleobase (a purine derivative). It is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The three others are guanine, cytosine and thymine. Its derivatives have a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate (ATP) and the cofactors nicotinamide adenine dinucleotide (NAD), flavin adenine dinucleotide (FAD) and Coenzyme A. It also has functions in protein synthesis and as a chemical component of DNA and RNA. The shape of adenine is complementary to either thymine in DNA or uracil in RNA. The adjacent image shows pure adenine, as an independent molecule. When connected into DNA, a covalent bond is formed between deoxyribose sugar and the bottom left nitrogen (thereby removing the existing hydrogen atom). The remaining structure is called an ''adenine residue'', as part of a larger molecule. Adenosine is ad ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphodiester Bond
In chemistry, a phosphodiester bond occurs when exactly two of the hydroxyl groups () in phosphoric acid react with hydroxyl groups on other molecules to form two ester bonds. The "bond" involves this linkage . Discussion of phosphodiesters is dominated by their prevalence in DNA and RNA, but phosphodiesters occur in other biomolecules, e.g. acyl carrier proteins. Phosphodiester bonds make up the backbones of DNA and RNA. The phosphate is attached to the 5' carbon. The 3' carbon of one sugar is bonded to the 5' phosphate of the adjacent sugar. Specifically, the phosphodiester bond links the 3' carbon atom of one sugar molecule and the 5' carbon atom of another (hence the name, 3', 5' phosphodiester linkage). These saccharide groups are derived from deoxyribose in DNA and ribose in RNA. Phosphodiesters are negatively charged at pH 7. Repulsion between these negative charges influences the conformation of the polynucleic acids. The negative charge attracts histones, metal c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribonuclease
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-specific RN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver. Nucleotides are composed of three subunit molecules: a nucleobase, a five-carbon sugar (ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. The four nucleobases in DNA are guanine, adenine, cytosine and thymine; in RNA, uracil is used in place of thymine. Nucleotides also play a central role in metabolism at a fundamental, cellular level. They provide chemical energy—in the form of the nucleoside triphosphates, adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP) and uridine triphosphate (UTP)—throughout the cell for the many cellular func ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribose
Ribose is a simple sugar and carbohydrate with molecular formula C5H10O5 and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally-occurring form, , is a component of the ribonucleotides from which RNA is built, and so this compound is necessary for coding, decoding, regulation and expression of genes. It has a structural analog, deoxyribose, which is a similarly essential component of DNA. is an unnatural sugar that was first prepared by Emil Fischer and Oscar Piloty in 1891. It was not until 1909 that Phoebus Levene and Walter Jacobs recognised that was a natural product, the enantiomer of Fischer and Piloty's product, and an essential component of nucleic acids. Fischer chose the name "ribose" as it is a partial rearrangement of the name of another sugar, arabinose, of which ribose is an epimer at the 2' carbon; both names also relate to gum arabic, from which arabinose was first isolated and from which they prepared . Like most sugars, ribose exists ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
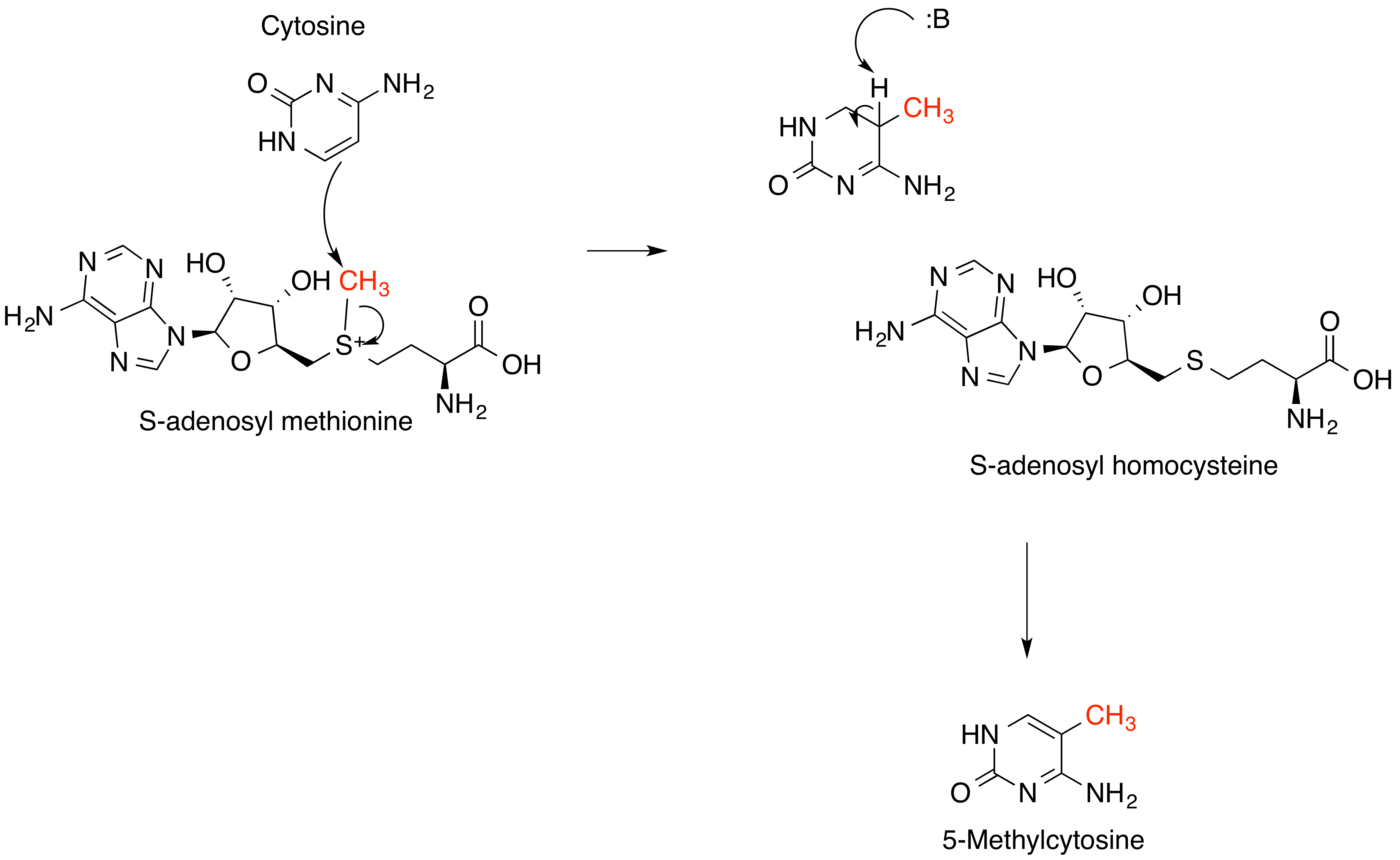
S-adenosyl Methionine
''S''-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM is produced and consumed in the liver. More than 40 methyl transfers from SAM are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM was first discovered by Giulio Cantoni in 1952. In bacteria, SAM is bound by the SAM riboswitch, which regulates genes involved in methionine or cysteine biosynthesis. In eukaryotic cells, SAM serves as a regulator of a variety of processes including DNA, tRNA, and rRNA methylation; immune response; amino acid metabolism; transsulfuration; and more. In plants, SAM is crucial to the biosynthesis of ethylene, an important plant hormone and sig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MRNA (guanine-N7-)-methyltransferase
In enzymology, a mRNA (guanine-N7-)-methyltransferase also known as mRNA cap guanine-N7 methyltransferase is an enzyme that catalyzes the chemical reaction :''S''-adenosyl-L-methionine + G(5')pppR-RNA \rightleftharpoons ''S''-adenosyl-L-homocysteine + m7G(5')pppR-RNA (mRNA containing an N7-methylguanine cap) Thus, the two substrates of this enzyme are S-adenosyl methionine and G(5')pppR-RNA, whereas its two products are S-adenosylhomocysteine and m7G(5')pppR-RNA. This enzyme belongs to the family of transferases, specifically those transferring one-carbon group methyltransferases. In humans, mRNA cap guanine-N7 methyltransferase is encoded by the ''RNMT'' gene. Nomenclature The systematic name of this enzyme class is S-adenosyl-L-methionine:mRNA (guanine-N7-)-methyltransferase. Other names in common use include: * messenger ribonucleate guanine 7-methyltransferase, * guanine-7-methyltransferase, * messenger RNA guanine 7-methyltransferase, and * S-adenosyl-L-methioni ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Guanine
Guanine () ( symbol G or Gua) is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine (uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside is called guanosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with conjugated double bonds. This unsaturated arrangement means the bicyclic molecule is planar. Properties Guanine, along with adenine and cytosine, is present in both DNA and RNA, whereas thymine is usually seen only in DNA, and uracil only in RNA. Guanine has two tautomeric forms, the major keto form (see figures) and rare enol form. It binds to cytosine through three hydrogen bonds. In cytosine, the amino group acts as the hydrogen bond donor and the C-2 carbonyl and the N-3 amine as the hydrogen-bond acceptors. Guanine has the C-6 carbonyl group that acts as the hydrogen bond acceptor, while a group at N ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Guanosine Triphosphate
Guanosine-5'-triphosphate (GTP) is a purine nucleoside triphosphate. It is one of the building blocks needed for the synthesis of RNA during the transcription process. Its structure is similar to that of the guanosine nucleoside, the only difference being that nucleotides like GTP have phosphates on their ribose sugar. GTP has the guanine nucleobase attached to the 1' carbon of the ribose and it has the triphosphate moiety attached to ribose's 5' carbon. It also has the role of a source of energy or an activator of substrates in metabolic reactions, like that of ATP, but more specific. It is used as a source of energy for protein synthesis and gluconeogenesis. GTP is essential to signal transduction, in particular with G-proteins, in second-messenger mechanisms where it is converted to guanosine diphosphate (GDP) through the action of GTPases. Uses Energy transfer GTP is involved in energy transfer within the cell. For instance, a GTP molecule is generated by one of the enz ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |