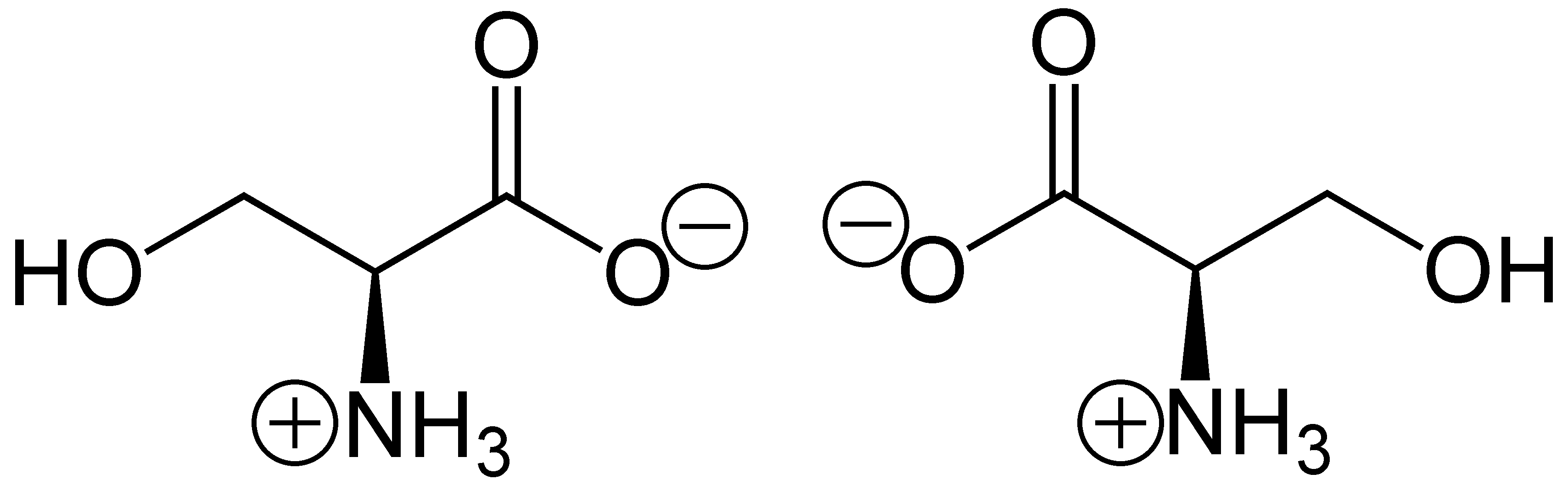|
Pasha (protein)
The microprocessor complex subunit DGCR8 ''(DiGeorge syndrome critical region 8)'' is a protein that in humans is encoded by the gene. In other animals, particularly the common model organisms ''Drosophila melanogaster'' and ''Caenorhabditis elegans'', the protein is known as ''Pasha'' (partner of Drosha). It is a required component of the RNA interference pathway. Function The subunit DGCR8 is localized to the cell nucleus and is required for microRNA (miRNA) processing. It binds to the other subunit Drosha, an RNase III enzyme, to form the microprocessor complex that cleaves a primary transcript known as pri-miRNA to a characteristic stem-loop structure known as a pre-miRNA, which is then further processed to miRNA fragments by the enzyme Dicer. DGCR8 contains an RNA-binding domain and is thought to bind pri-miRNA to stabilize it for processing by Drosha. DGCR8 is also required for some types of DNA repair. Removal of UV-induced DNA photoproducts, during transcription co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DiGeorge Syndrome
DiGeorge syndrome, also known as 22q11.2 deletion syndrome, is a syndrome caused by a microdeletion on the long arm of chromosome 22. While the symptoms can vary, they often include congenital heart problems, specific facial features, frequent infections, developmental delay, learning problems and cleft palate. Associated conditions include kidney problems, schizophrenia, hearing loss and autoimmune disorders such as rheumatoid arthritis or Graves' disease. DiGeorge syndrome is typically due to the deletion of 30 to 40 genes in the middle of chromosome 22 at a location known as ''22q11.2''. About 90% of cases occur due to a new mutation during early development, while 10% are inherited from a person's parents. It is autosomal dominant, meaning that only one affected chromosome is needed for the condition to occur. Diagnosis is suspected based on the symptoms and confirmed by genetic testing. Although there is no cure, treatment can improve symptoms. This often includes a m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogen Peroxide
Hydrogen peroxide is a chemical compound with the formula . In its pure form, it is a very pale blue liquid that is slightly more viscous than water. It is used as an oxidizer, bleaching agent, and antiseptic, usually as a dilute solution (3%–6% by weight) in water for consumer use, and in higher concentrations for industrial use. Concentrated hydrogen peroxide, or " high-test peroxide", decomposes explosively when heated and has been used as a propellant in rocketry. Hydrogen peroxide is a reactive oxygen species and the simplest peroxide, a compound having an oxygen–oxygen single bond. It decomposes slowly when exposed to light, and rapidly in the presence of organic or reactive compounds. It is typically stored with a stabilizer in a weakly acidic solution in a dark bottle to block light. Hydrogen peroxide is found in biological systems including the human body. Enzymes that use or decompose hydrogen peroxide are classified as peroxidases. Properties The boiling poi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L-stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure was estab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotide Excision Repair
Nucleotide excision repair is a DNA repair mechanism. DNA damage occurs constantly because of chemicals (e.g. intercalating agents), radiation and other mutagens. Three excision repair pathways exist to repair single stranded DNA damage: Nucleotide excision repair (NER), base excision repair (BER), and DNA mismatch repair (MMR). While the BER pathway can recognize specific non-bulky lesions in DNA, it can correct only damaged bases that are removed by specific glycosylases. Similarly, the MMR pathway only targets mismatched Watson-Crick base pairs. Nucleotide excision repair (NER) is a particularly important excision mechanism that removes DNA damage induced by ultraviolet light (UV). UV DNA damage results in bulky DNA adducts - these adducts are mostly thymine dimers and 6,4-photoproducts. Recognition of the damage leads to removal of a short single-stranded DNA segment that contains the lesion. The undamaged single-stranded DNA remains and DNA polymerase uses it as a templa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyrimidine Dimer
Pyrimidine dimers are molecular lesions formed from thymine or cytosine bases in DNA via photochemical reactions, commonly associated with direct DNA damage. Ultraviolet light (UV; particularly UVB) induces the formation of covalent linkages between consecutive bases along the nucleotide chain in the vicinity of their carbon–carbon double bonds. The dimerization reaction can also occur among pyrimidine bases in dsRNA (double-stranded RNA)—uracil or cytosine. Two common UV products are cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts. These premutagenic lesions alter the structure and possibly the base-pairing. Up to 50–100 such reactions per second might occur in a skin cell during exposure to sunlight, but are usually corrected within seconds by photolyase reactivation or nucleotide excision repair. Uncorrected lesions can inhibit polymerases, cause misreading during transcription or replication, or lead to arrest of replication. It causes sunburn and it tri ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dicer
Dicer, also known as endoribonuclease Dicer or helicase with RNase motif, is an enzyme that in humans is encoded by the gene. Being part of the RNase III family, Dicer cleaves double-stranded RNA (dsRNA) and pre-microRNA (pre-miRNA) into short double-stranded RNA fragments called small interfering RNA and microRNA, respectively. These fragments are approximately 20–25 base pairs long with a two-base overhang on the 3′-end. Dicer facilitates the activation of the RNA-induced silencing complex (RISC), which is essential for RNA interference. RISC has a catalytic component Argonaute, which is an endonuclease capable of degrading messenger RNA (mRNA). Discovery Dicer was given its name in 2001 by Stony Brook PhD student Emily Bernstein while conducting research in Gregory Hannon's lab at Cold Spring Harbor Laboratory. Bernstein sought to discover the enzyme responsible for generating small RNA fragments from double-stranded RNA. Dicer's ability to generate around 22 nucl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many RNA secondary structures. As an important secondary structure of RNA, it can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA binding proteins, and serve as a substrate for enzymatic reactions. Formation and stability The formation of a stem-loop structure is dependent on the stability of the resulting helix and loop regions. The first prerequisite is the presence of a sequence that can fold back on itself to form a paired double helix. The stability of this helix is determined by its length, the number of mismatches or bulges it co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Primary Transcript
A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs are modified in preparation for translation. For example, a precursor mRNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after processing. Pre-mRNA is synthesized from a DNA template in the cell nucleus by transcription. Pre-mRNA comprises the bulk of heterogeneous nuclear RNA (hnRNA). Once pre-mRNA has been completely processed, it is termed " mature messenger RNA", or simply "messenger RNA". The term hnRNA is often used as a synonym for pre-mRNA, although, in the strict sense, hnRNA may include nuclear RNA transcripts that do not end up as cytoplasmic mRNA. There are several steps contributing to the production of primary transcripts. All these steps involve a series of interactions to initiate and comp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microprocessor Complex
The microprocessor complex is a protein complex involved in the early stages of processing microRNA (miRNA) and RNA interference (RNAi) in animal cells. The complex is minimally composed of the ribonuclease enzyme Drosha and the dimeric RNA-binding protein DGCR8 (also known as Pasha in non-human animals), and cleaves primary miRNA substrates to pre-miRNA in the cell nucleus. Microprocessor is also the smaller of the two multi-protein complexes that contain human Drosha. Composition The microprocessor complex consists minimally of two proteins: Drosha, a ribonuclease III enzyme; and DGCR8, a double-stranded RNA binding protein. (DGCR8 is the name used in mammalian genetics, abbreviated from "DiGeorge syndrome critical region 8"; the homologous protein in model organisms such as flies and worms is called ''Pasha'', for ''Pa''rtner of Dro''sha''.) The stoichiometry of the minimal complex was at one point experimentally difficult to determine, but it has been demonstrated to be a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase III
Ribonuclease III (RNase III or RNase C)(BREND3.1.26.3 is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the ''pnp'' autoregulatory mechanism. Types of RNase III The RNase III superfamily is divided into four known classes: 1, 2, 3, and 4. Each class is defined by its domain structure.Liang Y-H, Lavoie M, Comeau M-A, Elela SA, Ji X. Structure of a Eukaryotic RNase III Post-Cleavage Complex Reveals a Double- Ruler Mechanism for Substrate Selection. Molecular cell. 2014;54(3):431-444. doi:10.1016/j.molcel.2014.03.006. Class 1 RNase III *Class 1 RNase III enzymes have a homodimeric structure whose function is to cleave dsRNA into multiple s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |