|
PLINK (genetic Tool-set)
PLINK is a free, commonly used, open-source whole-genome association analysis toolset designed by Shaun Purcell. The software is designed flexibly to perform a wide range of basic, large-scale genetic analyses. PLINK currently supports following functionalities: * data management; * basic statistics ( FST, missing data, tests of Hardy–Weinberg equilibrium, inbreeding coefficient, etc.); * Linkage disequilibrium (LD) calculation; * Identity by descent (IBD) and identity by state (IBS) matrix calculation; * population stratification, such as a Principal component analysis; * association analysis such as genome-wide association study In genomics, a genome-wide association study (GWA study, or GWAS), also known as whole genome association study (WGA study, or WGAS), is an observational study of a genome-wide set of genetic variants in different individuals to see if any vari ... for both basic case/control studies and quantitative traits; *tests for epistasis Input and o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Whole Genome Association Study
In genomics, a genome-wide association study (GWA study, or GWAS), also known as whole genome association study (WGA study, or WGAS), is an observational study of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms. When applied to human data, GWA studies compare the DNA of participants having varying phenotypes for a particular trait or disease. These participants may be people with a disease (cases) and similar people without the disease (controls), or they may be people with different phenotypes for a particular trait, for example blood pressure. This approach is known as phenotype-first, in which the participants are classified first by their clinical manifestation(s), as opposed to genotype-first. Each per ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Shaun Purcell
Shaun M. Purcell is a British genetic epidemiologist and statistical geneticist. He is a senior associate member of the Broad Institute of MIT and Harvard and its Stanley Center for Psychiatric Research. He is also a faculty member at the Brigham and Women's Hospital Department of Psychiatry. Having studied psychology and statistics at Oxford University, Purcell overcame the death of his supervisor David Fulker one year into a behavioural genetics programme at the Institute of Psychiatry to complete his Ph.D. under Pak Sham. He developed the PLINK genetics software during postdoctoral work with Mark Daly at the Whitehead Institute. He is also known for his research on the genetic bases of mental disorders such as schizophrenia and bipolar disorder, largely undertaken during faculty appointments at Massachusetts General Hospital and Mount Sinai in Manhattan Manhattan (), known regionally as the City, is the most densely populated and geographically smallest of the five bor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fixation Index
The fixation index (FST) is a measure of population differentiation due to genetic structure. It is frequently estimated from genetic polymorphism data, such as single-nucleotide polymorphisms (SNP) or microsatellites. Developed as a special case of Wright's F-statistics, it is one of the most commonly used statistics in population genetics. Definition Two of the most commonly used definitions for FST at a given locus are based on 1) the variance of allele frequencies between populations, and on 2) the probability of Identity by descent. If \bar is the average frequency of an allele in the total population, \sigma^2_S is the variance in the frequency of the allele between different subpopulations, weighted by the sizes of the subpopulations, and \sigma^2_T is the variance of the allelic state in the total population, FST is defined as : F_ = \frac = \frac Wright's definition illustrates that FST measures the amount of genetic variance that can be explained by population stru ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Inbreeding Coefficient
The coefficient of inbreeding of an individual is the probability that two alleles at any locus in an individual are identical by descent from the common ancestor(s) of the two parents. The coefficient of inbreeding is: The probability that two alleles at a given locus are identical by descent. The coefficient of relationship is: The proportion Proportionality, proportion or proportional may refer to: Mathematics * Proportionality (mathematics), the property of two variables being in a multiplicative relation to a constant * Ratio, of one quantity to another, especially of a part compare ... of genes that are held in common by two individuals as a result of direct or collateral relationship. References Population genetics {{genetics-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Linkage Disequilibrium
In population genetics, linkage disequilibrium (LD) is the non-random association of alleles at different loci in a given population. Loci are said to be in linkage disequilibrium when the frequency of association of their different alleles is higher or lower than what would be expected if the loci were independent and associated randomly. Linkage disequilibrium is influenced by many factors, including selection, the rate of genetic recombination, mutation rate, genetic drift, the system of mating, population structure, and genetic linkage. As a result, the pattern of linkage disequilibrium in a genome is a powerful signal of the population genetic processes that are structuring it. In spite of its name, linkage disequilibrium may exist between alleles at different loci without any genetic linkage between them and independently of whether or not allele frequencies are in equilibrium (not changing with time). Furthermore, linkage disequilibrium is sometimes referred to as gamet ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Identity By Descent
A DNA segment is identical by state (IBS) in two or more individuals if they have identical nucleotide sequences in this segment. An IBS segment is identical by descent (IBD) in two or more individuals if they have inherited it from a common ancestor without recombination, that is, the segment has the same ancestral origin in these individuals. DNA segments that are IBD are IBS per definition, but segments that are not IBD can still be IBS due to the same mutations in different individuals or recombinations that do not alter the segment. Theory All individuals in a finite population are related if traced back long enough and will, therefore, share segments of their genomes IBD. During meiosis segments of IBD are broken up by recombination. Therefore, the expected length of an IBD segment depends on the number of generations since the most recent common ancestor at the locus of the segment. The length of IBD segments that result from a common ancestor ''n'' generations in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Identity By State
A DNA segment is identical by state (IBS) in two or more individuals if they have identical nucleotide sequences in this segment. An IBS segment is identical by descent (IBD) in two or more individuals if they have inherited it from a common ancestor without recombination, that is, the segment has the same ancestral origin in these individuals. DNA segments that are IBD are IBS per definition, but segments that are not IBD can still be IBS due to the same mutations in different individuals or recombinations that do not alter the segment. Theory All individuals in a finite population are related if traced back long enough and will, therefore, share segments of their genomes IBD. During meiosis segments of IBD are broken up by recombination. Therefore, the expected length of an IBD segment depends on the number of generations since the most recent common ancestor at the locus of the segment. The length of IBD segments that result from a common ancestor ''n'' generations in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Population Stratification
Population structure (also called genetic structure and population stratification) is the presence of a systematic difference in allele frequencies between subpopulations. In a randomly mating (or ''panmictic'') population, allele frequencies are expected to be roughly similar between groups. However, mating tends to be non-random to some degree, causing structure to arise. For example, a barrier like a river can separate two groups of the same species and make it difficult for potential mates to cross; if a mutation occurs, over many generations it can spread and become common in one subpopulation while being completely absent in the other. Genetic variants do not necessarily cause observable changes in organisms, but can be correlated by coincidence because of population structure—a variant that is common in a population that has a high rate of disease may erroneously be thought to cause the disease. For this reason, population structure is a common confounding variable in medi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Principal Component Analysis
Principal component analysis (PCA) is a popular technique for analyzing large datasets containing a high number of dimensions/features per observation, increasing the interpretability of data while preserving the maximum amount of information, and enabling the visualization of multidimensional data. Formally, PCA is a statistical technique for reducing the dimensionality of a dataset. This is accomplished by linearly transforming the data into a new coordinate system where (most of) the variation in the data can be described with fewer dimensions than the initial data. Many studies use the first two principal components in order to plot the data in two dimensions and to visually identify clusters of closely related data points. Principal component analysis has applications in many fields such as population genetics, microbiome studies, and atmospheric science. The principal components of a collection of points in a real coordinate space are a sequence of p unit vectors, where th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
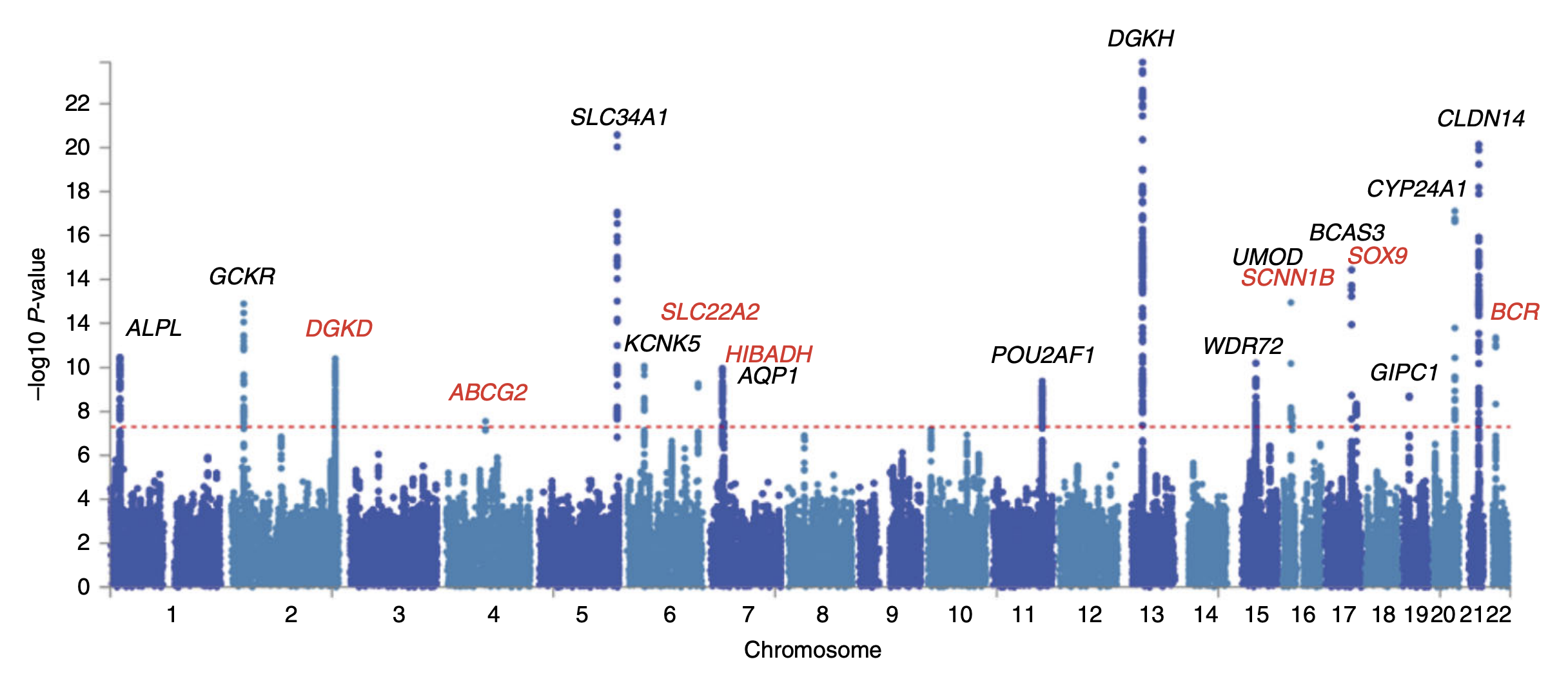
Genome-wide Association Study
In genomics, a genome-wide association study (GWA study, or GWAS), also known as whole genome association study (WGA study, or WGAS), is an observational study of a genome-wide set of Single-nucleotide polymorphism, genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms. When applied to human data, GWA studies compare the DNA of participants having varying phenotypes for a particular trait or disease. These participants may be people with a disease (cases) and similar people without the disease (controls), or they may be people with different phenotypes for a particular trait, for example blood pressure. This approach is known as phenotype-first, in which the participants are classified first by their clinical manifestation(s), as oppose ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
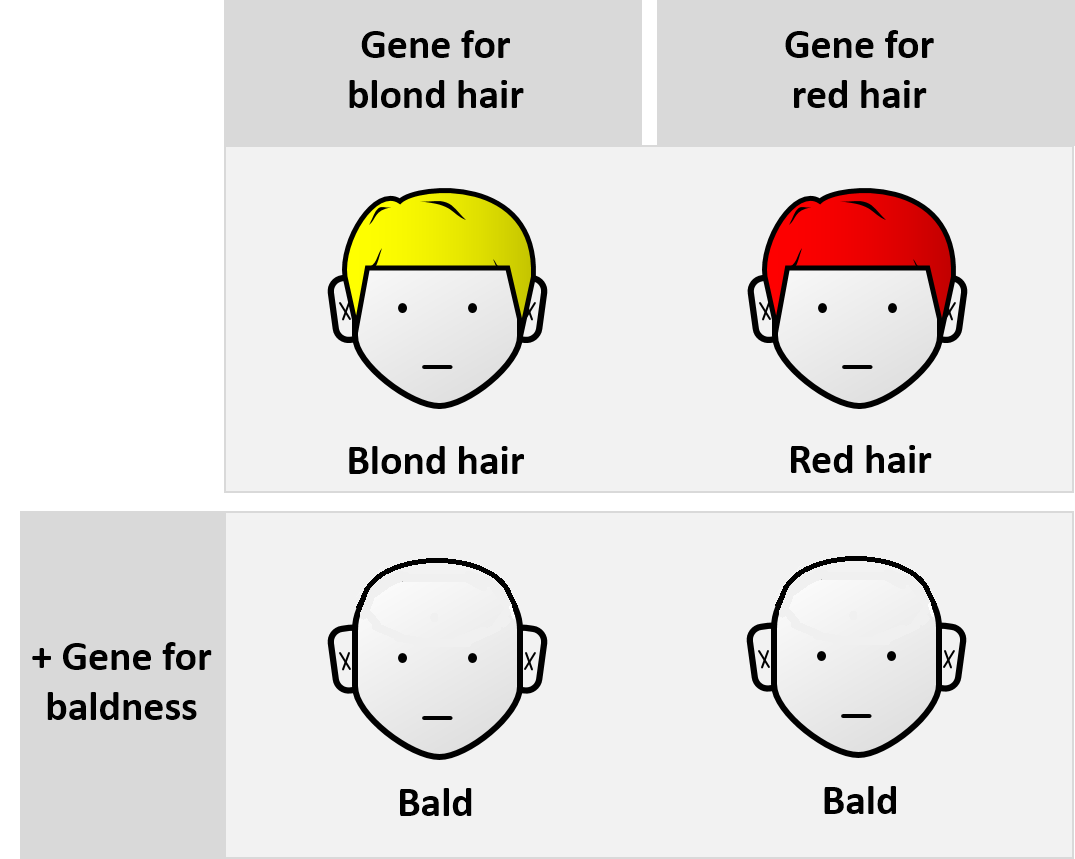
Epistasis
Epistasis is a phenomenon in genetics in which the effect of a gene mutation is dependent on the presence or absence of mutations in one or more other genes, respectively termed modifier genes. In other words, the effect of the mutation is dependent on the genetic background in which it appears. Epistatic mutations therefore have different effects on their own than when they occur together. Originally, the term ''epistasis'' specifically meant that the effect of a gene variant is masked by that of a different gene. The concept of ''epistasis'' originated in genetics in 1907 but is now used in biochemistry, computational biology and evolutionary biology. The phenomenon arises due to interactions, either between genes (such as mutations also being needed in regulators of gene expression) or within them (multiple mutations being needed before the gene loses function), leading to non-linear effects. Epistasis has a great influence on the shape of evolutionary landscapes, which l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |