|
PGLYRP4
Peptidoglycan recognition protein 4 (PGLYRP4, formerly PGRP-Iβ) is an antibacterial and anti-inflammatory innate immunity protein that in humans is encoded by the ''PGLYRP4'' gene. Discovery PGLYRP4 (formerly PGRP-Iβ), a member of a family of human Peptidoglycan Recognition Proteins (PGRPs), was discovered in 2001 by Roman Dziarski and coworkers who cloned and identified the genes for three human PGRPs, PGRP-L, PGRP-Iα, and PGRP-Iβ (named for long and intermediate size transcripts), and established that human genome codes for a family of 4 PGRPs: PGRP-S (short PGRP or PGRP-S) and PGRP-L, PGRP-Iα, and PGRP-Iβ. Subsequently, the Human Genome Organization Gene Nomenclature Committee changed the gene symbols of PGRP-S, PGRP-L, PGRP-Iα, and PGRP-Iβ to ''PGLYRP1'' (peptidoglycan recognition protein 1), ''PGLYRP2'' (peptidoglycan recognition protein 2), ''PGLYRP3'' (peptidoglycan recognition protein 3), and ''PGLYRP4'' (peptidoglycan recognition protein 4), respectively, and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptidoglycan Recognition Protein
Peptidoglycan recognition proteins (PGRPs) are a group of highly conserved pattern recognition receptors with at least one peptidoglycan recognition domain capable of recognizing the peptidoglycan component of the cell wall of bacteria. They are present in insects, mollusks, echinoderms and chordates. The mechanism of action of PGRPs varies between taxa. In insects, PGRPs kill bacteria indirectly by activating one of four unique effector pathways: prophenoloxidase cascade, Toll pathway, IMD pathway, and induction of phagocytosis. In mammals, PGRPs either kill bacteria directly by interacting with their cell wall or outer membrane, or hydrolyze peptidoglycan. They also modulate inflammation and microbiome and interact with host receptors. Discovery The first PGRP was discovered in 1996 by Masaaki Ashida and coworkers, who purified a 19 kDa protein present in the hemolymph and cuticle of a silkworm (''Bombyx mori''), and named it Peptidoglycan Recognition Protein, because it specif ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Roman Dziarski
Roman Dziarski (Polish pronunciation: IPA: /ˈrɔ.man//ˈd͡ʑar.ski/ born December 11, 1949) is a Polish-born American immunologist and microbiologist. He is best known for his research on innate immunity and bacterial peptidoglycan, for discovering the family of human peptidoglycan recognition proteins, which comprises PGLYRP1, PGLYRP2, PGLYRP3, and PGLYRP4, and for defining the functions of these proteins. Dziarski is currently Professor Emeritus of Microbiology and Immunology at Indiana University School of Medicine. Education From 1963 to 1967, Dziarski received his secondary education at Reytan High School (Polish: ''VI Liceum Ogólnokształcące im. Tadeusza Reytana'') in Warsaw, Poland, under the tutelage of the revered pedagogue, . From 1967 to 1972, Dziarski attended the University of Warsaw with a major in Biology and Microbiology, which he studied under . He received his Bachelor of Sciences (BS) degree in 1971, and Master of Science (MS) degree in 1972. His M ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptidoglycan Recognition Protein 3
Peptidoglycan recognition protein 3 (PGLYRP3, formerly PGRP-Iα) is an antibacterial and anti-inflammatory innate immunity protein that in humans is encoded by the ''PGLYRP3'' gene. Discovery PGLYRP3 (formerly PGRP-Iα), a member of a family of human Peptidoglycan Recognition Proteins (PGRPs), was discovered in 2001 by Roman Dziarski and coworkers who cloned and identified the genes for three human PGRPs, PGRP-L, PGRP-Iα, and PGRP-Iβ (named for long and intermediate size transcripts), and established that human genome codes for a family of 4 PGRPs: PGRP-S (short PGRP or PGRP-S) and PGRP-L, PGRP-Iα, and PGRP-Iβ. Subsequently, the Human Genome Organization Gene Nomenclature Committee changed the gene symbols of PGRP-S, PGRP-L, PGRP-Iα, and PGRP-Iβ to ''PGLYRP1'' (peptidoglycan recognition protein 1), ''PGLYRP2'' ( peptidoglycan recognition protein 2), ''PGLYRP3'' (peptidoglycan recognition protein 3), and ''PGLYRP4'' ( peptidoglycan recognition protein 4), respectively, a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptidoglycan Recognition Protein 2
Peptidoglycan recognition protein 2 (PGLYRP2) is an enzyme (EC 3.5.1.28), ''N''-acetylmuramoyl-L-alanine amidase (NAMLAA), that hydrolyzes bacterial cell wall peptidoglycan and is encoded by the ''PGLYRP2'' gene. Discovery The ''N''-acetylmuramoyl-L-alanine amidase enzymatic activity was first observed in human and mouse serum in 1981 by Branko Ladešić and coworkers. The enzyme (abbreviated NAMLAA) was then purified from human serum by this and other groups. The sequence of 15 N-terminal amino acids of NAMLAA was identified, but the cDNA for the protein was not cloned and the gene encoding NAMLAA was not known. In 2000, Dan Hultmark and coworkers discovered a family of 12 Peptidoglycan Recognition Protein (PGRP) genes in ''Drosophila'' ''melanogaster'' and by homology searches of available human and mouse sequences predicted the presence of long forms of human and mouse PGRPs, which they named PGRP-L by analogy to long forms of insect PGRPs. In 2001, Roman Dziarski and c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptidoglycan Recognition Protein 1
Peptidoglycan recognition protein 1, PGLYRP1, also known as TAG7, is an antibacterial and pro-inflammatory innate immunity protein that in humans is encoded by the ''PGLYRP1'' gene. Discovery PGLYRP1 was discovered independently by two laboratories in 1998. Håkan Steiner and coworkers, using a differential display screen, identified and cloned Peptidoglycan Recognition Protein (PGRP) in a moth (''Trichoplusia ni'') and based on this sequence discovered and cloned mouse and human PGRP orthologs. Sergei Kiselev and coworkers discovered and cloned a protein from a mouse adenocarcinoma with the same sequence as mouse PGRP, which they named Tag7. Human PGRP was a founding member of a family of four PGRP genes found in humans that were named PGRP-S, PGRP-L, PGRP-Iα, and PGRP-Iβ (for short, long, and intermediate size transcripts, by analogy to insect PGRPs). Their gene symbols were subsequently changed to ''PGLYRP1'' (peptidoglycan recognition protein 1), ''PGLYRP2'' ( peptidogly ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epithelium
Epithelium or epithelial tissue is one of the four basic types of animal tissue, along with connective tissue, muscle tissue and nervous tissue. It is a thin, continuous, protective layer of compactly packed cells with a little intercellular matrix. Epithelial tissues line the outer surfaces of organs and blood vessels throughout the body, as well as the inner surfaces of cavities in many internal organs. An example is the epidermis, the outermost layer of the skin. There are three principal shapes of epithelial cell: squamous (scaly), columnar, and cuboidal. These can be arranged in a singular layer of cells as simple epithelium, either squamous, columnar, or cuboidal, or in layers of two or more cells deep as stratified (layered), or ''compound'', either squamous, columnar or cuboidal. In some tissues, a layer of columnar cells may appear to be stratified due to the placement of the nuclei. This sort of tissue is called pseudostratified. All glands are made up of epithe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Brain
A brain is an organ that serves as the center of the nervous system in all vertebrate and most invertebrate animals. It is located in the head, usually close to the sensory organs for senses such as vision. It is the most complex organ in a vertebrate's body. In a human, the cerebral cortex contains approximately 14–16 billion neurons, and the estimated number of neurons in the cerebellum is 55–70 billion. Each neuron is connected by synapses to several thousand other neurons. These neurons typically communicate with one another by means of long fibers called axons, which carry trains of signal pulses called action potentials to distant parts of the brain or body targeting specific recipient cells. Physiologically, brains exert centralized control over a body's other organs. They act on the rest of the body both by generating patterns of muscle activity and by driving the secretion of chemicals called hormones. This centralized control allows rapid and coordinated respon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microbiome
A microbiome () is the community of microorganisms that can usually be found living together in any given habitat. It was defined more precisely in 1988 by Whipps ''et al.'' as "a characteristic microbial community occupying a reasonably well-defined habitat which has distinct physio-chemical properties. The term thus not only refers to the microorganisms involved but also encompasses their theatre of activity". In 2020, an international panel of experts published the outcome of their discussions on the definition of the microbiome. They proposed a definition of the microbiome based on a revival of the "compact, clear, and comprehensive description of the term" as originally provided by Whipps ''et al.'', but supplemented with two explanatory paragraphs. The first explanatory paragraph pronounces the dynamic character of the microbiome, and the second explanatory paragraph clearly separates the term ''microbiota'' from the term ''microbiome''. The microbiota consists of all ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disulfide
In biochemistry, a disulfide (or disulphide in British English) refers to a functional group with the structure . The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. ''Persulfide'' usually refers to compounds. In inorganic chemistry disulfide usually refers to the corresponding anion (−S−S−). Organic disulfides Symmetrical disulfides are compounds of the formula . Most disulfides encountered in organo sulfur chemistry are symmetrical disulfides. Unsymmetrical disulfides (also called heterodisulfides) are compounds of the formula . They are less common in organic chemistry, but most disulfides in nature are unsymmetrical. Properties The disulfide bonds are strong, with a typical bond dissociation energy of 60 kcal/mol (251&nbs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-acetylmuramoyl-L-alanine Amidase
In enzymology, a N-acetylmuramoyl-L-alanine amidase () is an enzyme that catalysis, catalyzes a chemical reaction that cleaves the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell wall, cell-wall glycopeptides. This enzyme belongs to the family of hydrolases, specifically those acting on carbon-nitrogen bonds other than peptide bonds in linear amides. The List of enzymes, systematic name of this enzyme class is peptidoglycan amidohydrolase. Other names in common use include acetylmuramyl-L-alanine amidase, N-acetylmuramyl-L-alanine amidase, N-acylmuramyl-L-alanine amidase, acetylmuramoyl-alanine amidase, N-acetylmuramic acid L-alanine amidase, acetylmuramyl-alanine amidase, N-acetylmuramylalanine amidase, N-acetylmuramoyl-L-alanine amidase type I, and N-acetylmuramoyl-L-alanine amidase type II. This enzyme participates in peptidoglycan biosynthesis. Autolysins and some phage lysins are examples of N-acetylmuramoyl-L-alanine amidases. See also * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human PGLYRP4 Gene, CDNA, And Protein
Humans (''Homo sapiens'') are the most abundant and widespread species of primate, characterized by bipedalism and exceptional cognitive skills due to a large and complex brain. This has enabled the development of advanced tools, culture, and language. Humans are highly social and tend to live in complex social structures composed of many cooperating and competing groups, from families and kinship networks to political states. Social interactions between humans have established a wide variety of values, social norms, and rituals, which bolster human society. Its intelligence and its desire to understand and influence the environment and to explain and manipulate phenomena have motivated humanity's development of science, philosophy, mythology, religion, and other fields of study. Although some scientists equate the term ''humans'' with all members of the genus ''Homo'', in common usage, it generally refers to ''Homo sapiens'', the only extant member. Anatomically modern ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
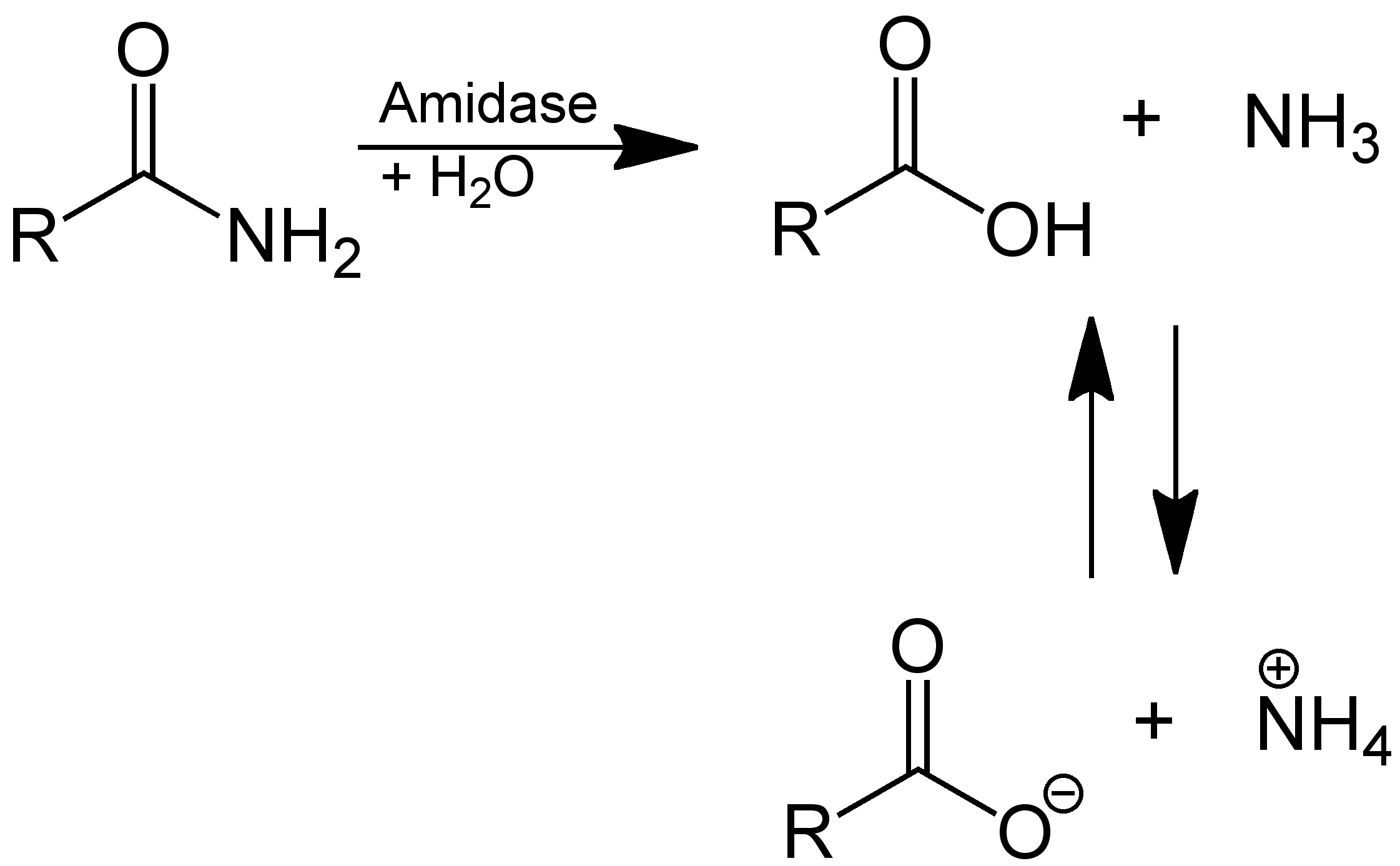
Amidase
In enzymology, an amidase (, ''acylamidase'', ''acylase (misleading)'', ''amidohydrolase (ambiguous)'', ''deaminase (ambiguous)'', ''fatty acylamidase'', ''N-acetylaminohydrolase (ambiguous)'') is an enzyme that catalyzes the hydrolysis of an amide: Thus, the two substrates of this enzyme are an amide and H2O, whereas its two products are monocarboxylate and NH3. This enzyme belongs to the family of hydrolases, those acting on carbon-nitrogen bonds other than peptide bonds, specifically in linear amides. The systematic name of this enzyme class is acylamide amidohydrolase. Other names in common use include acylamidase, acylase, amidohydrolase, deaminase, fatty acylamidase, and N-acetylaminohydrolase. This enzyme participates in 6 metabolic pathways: urea cycle and metabolism of amino groups, phenylalanine metabolism, tryptophan metabolism, cyanoamino acid metabolism, benzoate degradation via coa ligation, and styrene degradation. Amidases contain a conserved stretch o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |