|
OpenMS
OpenMS is an open-source project for data analysis and processing in mass spectrometry and is released under the 3-clause BSD licence. It supports most common operating systems including Microsoft Windows, MacOS and Linux. OpenMS has tools for analysis of proteomics data, providing algorithms for signal processing, feature finding (including de-isotoping), visualization in 1D (spectra or chromatogram level), 2D and 3D, map mapping and peptide identification. It supports label-free and isotopic-label based quantification (such as iTRAQ and TMT and SILAC). OpenMS also supports metabolomics workflows and targeted analysis of DIA/SWATH data. Furthermore, OpenMS provides tools for the analysis of cross linking data, including protein-protein, protein-RNA and protein-DNA cross linking. Lastly, OpenMS provides tools for analysis of RNA mass spectrometry data. History OpenMS was originally released in 2007 in version 1.0 and was described in two articles published in Bioinformati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mass Spectrometry Software
Mass spectrometry software is software used for data acquisition, analysis, or representation in mass spectrometry. Proteomics software In protein mass spectrometry, tandem mass spectrometry (also known as MS/MS or MS2) experiments are used for protein/peptide identification. Peptide identification algorithms fall into two broad classes: database search and ''de novo'' search. The former search takes place against a database containing all amino acid sequences assumed to be present in the analyzed sample, whereas the latter infers peptide sequences without knowledge of genomic data. Database search algorithms De novo sequencing algorithms De novo peptide sequencing algorithms are based, in general, on the approach proposed in Bartels ''et al.'' (1990). Homology searching algorithms MS/MS peptide quantification Other software See also * Mass spectrometry data format: for a list of mass spectrometry data viewers and format converters. * List of prote ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Trans-Proteomic Pipeline
The Trans-Proteomic Pipeline (TPP) is an open-source data analysis software for proteomics developed at the Institute for Systems Biology (ISB) by the Ruedi Aebersold group under the Seattle Proteome Center. The TPP includes PeptideProphet, ProteinProphet, ASAPRatio, XPRESS and Libra. Software Components Probability Assignment and Validation PeptideProphet performs statistical validation of peptide-spectra-matches (PSM) using the results of search engines by estimating a false discovery rate (FDR) on PSM level. The initial PeptideProphet used a fit of a Gaussian distribution for the correct identifications and a fit of a gamma distribution for the incorrect identification. A later modification of the program allowed the usage of a target-decoy approach, using either a variable component mixture model or a semi-parametric mixture model. In the PeptideProphet, specifying a decoy tag will use the variable component mixture model while selecting a non-parametric model will use ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ProteoWizard
ProteoWizard is a set of open-source, cross-platform tools and libraries for proteomics data analyses. It provides a framework for unified mass spectrometry data file access and performs standard chemistry and LCMS dataset computations. Specifically, it is able to read many of the vendor-specific, proprietary formats and converting the data into an open data format. On the application level, the software provides executables for data conversion (msConvert, msConvertGUI and idConvert), data visualization (msPicture and seeMS), data access (msAccess, msCat, idCat and msPicture), data analysis (peekaboo and msPrefix14) and basic proteomics utilities (chainsaw). In addition, the project also hosts the Skyline software which helps to create, acquire and analyze targeted proteomics experiments such as SRM experiments. The main contributors to the project are the Tabb, MacCoss and Mallick research labs as well as Insilicos. See also * OpenMS * Trans-Proteomic Pipeline * Mass spectro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics (journal)
''Bioinformatics'' (sometimes called ''Bioinformatics Oxford journal'') is a biweekly peer-reviewed scientific journal covering research and software in bioinformatics and computational biology. It is the official journal of the International Society for Computational Biology (ISCB), together with ''PLOS Computational Biology''. Authors can pay extra for open access and are allowed to self-archive after 1 year. The journal was established as ''Computer Applications in the Biosciences'' (''CABIOS'') in 1985. The founding editor-in-chief was Robert J. Beynon. In 1998, the journal obtained its current name and established an online version of the journal. It is published by Oxford University Press and, as of 2014, the editors-in-chief are Alfonso Valencia and Janet Kelso. Previous editors include Chris Sander, Gary Stormo, Christos Ouzounis, Martin Bishop, and Alex Bateman. In 2014, these five editors were appointed the first Honorary Editors of ''Bioinformatics''. According to t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Git (software)
Git () is a distributed version control system: tracking changes in any set of files, usually used for coordinating work among programmers collaboratively developing source code during software development. Its goals include speed, data integrity, and support for distributed, non-linear workflows (thousands of parallel branches running on different systems). "So I'm writing some scripts to try to track things a whole lot faster." Git was originally authored by Linus Torvalds in 2005 for development of the Linux kernel, with other kernel developers contributing to its initial development. Since 2005, Junio Hamano has been the core maintainer. As with most other distributed version control systems, and unlike most client–server systems, every Git directory on every computer is a full-fledged repository with complete history and full version-tracking abilities, independent of network access or a central server. Git is free and open-source software distributed under the GP ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
KNIME
KNIME (), the Konstanz Information Miner, is a free and open-source data analytics, reporting and integration platform. KNIME integrates various components for machine learning and data mining through its modular data pipelining "Building Blocks of Analytics" concept. A graphical user interface and use of JDBC allows assembly of nodes blending different data sources, including preprocessing ( ETL: Extraction, Transformation, Loading), for modeling, data analysis and visualization without, or with only minimal, programming. Since 2006, KNIME has been used in pharmaceutical research, it also used in other areas such as CRM customer data analysis, business intelligence, text mining and financial data analysis. Recently attempts were made to use KNIME as robotic process automation (RPA) tool. KNIME's headquarters are based in Zurich, with additional offices in Konstanz, Berlin, and Austin (USA). History The Development of KNIME was started January 2004 by a team of software engin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
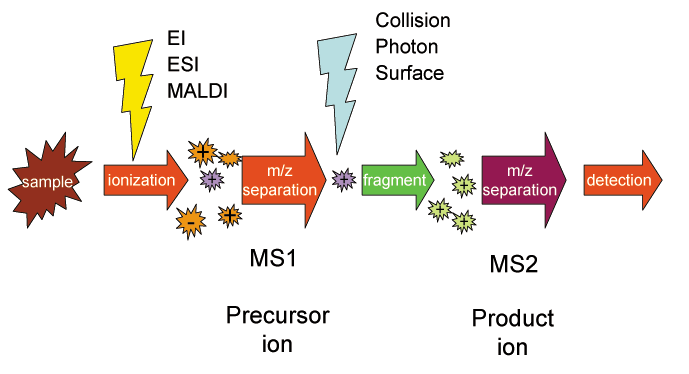
Selected Reaction Monitoring
Selected reaction monitoring (SRM), also called Multiple reaction monitoring, (MRM), is a method used in tandem mass spectrometry in which an ion of a particular mass is selected in the first stage of a tandem mass spectrometer and an ion product of a fragmentation reaction of the precursor ions is selected in the second mass spectrometer stage for detection. Variants A general case of SRM can be represented by :ABCD^+ \to AB + CD^+ where the precursor ion ABCD+ is selected by the first stage of mass spectrometry (MS1), dissociates into molecule AB and product ion CD+, and the latter is selected by the second stage of mass spectrometry (MS2) and detected. The precursor and product ion pair is called a SRM "transition." Consecutive reaction monitoring (CRM) is the serial application of three or more stages of mass spectrometry to SRM, represented in a simple case by :ABCD^+ \to AB + CD^+ \to C + D^+ where ABCD+ is selected by MS1, dissociates into molecule AB and ion CD+. Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ETH Zurich
(colloquially) , former_name = eidgenössische polytechnische Schule , image = ETHZ.JPG , image_size = , established = , type = Public , budget = CHF 1.896 billion (2021) , rector = Günther Dissertori , president = Joël Mesot , academic_staff = 6,612 (including doctoral students, excluding 527 professors of all ranks, 34% female, 65% foreign nationals) (full-time equivalents 2021) , administrative_staff = 3,106 (40% female, 19% foreign nationals, full-time equivalents 2021) , students = 24,534 (headcount 2021, 33.3% female, 37% foreign nationals) , undergrad = 10,642 , postgrad = 8,299 , doctoral = 4,460 , other = 1,133 , address = Rämistrasse 101CH-8092 ZürichSwitzerland , city = Zürich , coor = , campus = Urban , language = German, English (Masters and upwards, sometimes Bachelor) , affiliations = CESAER, EUA, GlobalTech, IARU, IDEA League, UNITECH , website ethz.ch, colors = Black and White , logo = ETH Zürich Logo black.svg ETH Zür ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ruedi Aebersold
Rudolf Aebersold (better known as Ruedi Aebersold born September 12, 1954 ) is a Swiss biologist, regarded as a pioneer in the fields of proteomics and systems biology. He has primarily researched techniques for measuring proteins in complex samples, in many cases via mass spectrometry. Ruedi Aebersold is a professor of Systems biology at the Institute of Molecular Systems Biology (IMSB) in ETH Zurich. He was one of the founders of the Institute for Systems Biology in Seattle, Washington, where he previously had a research group. Ruedi Aebersold is known for the development and application of targeted proteomics techniques in the field of biomedical research, in order to understand the function, interaction and localization of each protein in the cell and its changes in disease states. To this end, Ruedi Abersold has made significant contributions in the development and application of targeted proteomics methods, including selected reaction monitoring and data-independent acquis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
University Of Tübingen
The University of Tübingen, officially the Eberhard Karl University of Tübingen (german: Eberhard Karls Universität Tübingen; la, Universitas Eberhardina Carolina), is a public research university located in the city of Tübingen, Baden-Württemberg, Germany. The University of Tübingen is one of eleven German Excellence Universities. The University of Tübingen is especially known as a centre for the study of plant biology, medicine, law, archeology, ancient cultures, philosophy, theology, and religious studies as well as more recently as center of excellence for artificial intelligence. The university's noted alumni include presidents, EU Commissioners, and judges of the Federal Constitutional Court. The university is associated with eleven Nobel laureates, especially in the fields of medicine and chemistry. History The University of Tübingen was founded in 1477 by Count Eberhard V (Eberhard im Bart, 1445–1496), later the first Duke of Württemberg, a civic a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Free University Of Berlin
The Free University of Berlin (, often abbreviated as FU Berlin or simply FU) is a public research university in Berlin, Germany. It is consistently ranked among Germany's best universities, with particular strengths in political science and the humanities. It is recognised as a leading university in international university rankings. The Free University of Berlin was founded in West Berlin in 1948 with American support during the early Cold War period as a Western continuation of the Friedrich Wilhelm University, or the University of Berlin, whose traditions and faculty members it retained. The Friedrich Wilhelm University (which was renamed the Humboldt University), being in East Berlin, faced strong communist repression; the Free University's name referred to West Berlin's status as part of the Western Free World, in contrast to communist-controlled East Berlin. In 2008, as part of a joint effort, the Free University of Berlin, along with the Hertie School of Govern ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nature Methods
''Nature Methods'' is a monthly peer-reviewed scientific journal covering new scientific techniques. It was established in 2004 and is published by Springer Nature under the Nature Portfolio. Like other ''Nature'' journals, there is no external editorial board and editorial decisions are made by an in-house team, although peer review by external experts forms a part of the review process. Every year, the journal highlights a field, approach, or technique that has enabled recent major advances in life sciences research as the "Method of the Year". According to the ''Journal Citation Reports'', the journal had a 2021 impact factor The impact factor (IF) or journal impact factor (JIF) of an academic journal is a scientometric index calculated by Clarivate that reflects the yearly mean number of citations of articles published in the last two years in a given journal, as ... of 47.99, ranking it first in the category "Biochemical Research Methods". References External link ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |