|
Molecular Biophysics
Molecular biophysics is a rapidly evolving interdisciplinary area of research that combines concepts in physics, chemistry, engineering, mathematics and biology. It seeks to understand biomolecular systems and explain biological function in terms of molecular structure, structural organization, and dynamic behaviour at various levels of complexity (from single molecules to supramolecular structures, viruses and small living systems). This discipline covers topics such as the measurement of molecular forces, molecular associations, allosteric interactions, Brownian motion, and cable theory. Additional areas of study can be found on Outline of Biophysics. The discipline has required development of specialized equipment and procedures capable of imaging and manipulating minute living structures, as well as novel experimental approaches. Overview Molecular biophysics typically addresses biological questions similar to those in biochemistry and molecular biology, seeking to find ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Translation
In biology, translation is the process in living cells in which proteins are produced using RNA molecules as templates. The generated protein is a sequence of amino acids. This sequence is determined by the sequence of nucleotides in the RNA. The nucleotides are considered three at a time. Each such triple results in the addition of one specific amino acid to the protein being generated. The matching from nucleotide triple to amino acid is called the genetic code. The translation is performed by a large complex of functional RNA and proteins called ribosomes. The entire process is called gene expression. In translation, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later folds into an active protein and performs its functions in the cell. The polypeptide can also start folding during protein synthesis. The ribosome facilitates decoding by inducing the binding of complementary tra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electron Microscopy
An electron microscope is a microscope that uses a beam of electrons as a source of illumination. It uses electron optics that are analogous to the glass lenses of an optical light microscope to control the electron beam, for instance focusing it to produce magnified images or electron diffraction patterns. As the wavelength of an electron can be up to 100,000 times smaller than that of visible light, electron microscopes have a much higher resolution of about 0.1 nm, which compares to about 200 nm for light microscopes. ''Electron microscope'' may refer to: * Transmission electron microscope (TEM) where swift electrons go through a thin sample * Scanning transmission electron microscope (STEM) which is similar to TEM with a scanned electron probe * Scanning electron microscope (SEM) which is similar to STEM, but with thick samples * Electron microprobe similar to a SEM, but more for chemical analysis * Low-energy electron microscope (LEEM), used to image surfaces * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Optical Tweezers
Optical tweezers (originally called single-beam gradient force trap) are scientific instruments that use a highly focused laser beam to hold and move microscopic and sub-microscopic objects like atoms, nanoparticles and droplets, in a manner similar to tweezers. If the object is held in air or vacuum without additional support, it can be called optical levitation. The laser light provides an attractive or repulsive force (typically on the order of piconewtons), depending on the relative refractive index between particle and surrounding medium. Levitation is possible if the force of the light counters the force of gravity. The trapped particles are usually micron-sized, or even smaller. Dielectric and absorbing particles can be trapped, too. Optical tweezers are used in biology and medicine (for example to grab and hold a single bacterium, a cell like a sperm cell or a blood cell, or a molecule like DNA), nanoengineering and nanochemistry (to study and build materials from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Circular Dichroism
Circular dichroism (CD) is dichroism involving circular polarization, circularly polarized light, i.e., the differential Absorption (electromagnetic radiation), absorption of left- and right-handed light. Left-hand circular (LHC) and right-hand circular (RHC) polarized light represent two possible spin angular momentum of light, spin angular momentum states for a photon, and so circular dichroism is also referred to as dichroism for spin angular momentum. This phenomenon was discovered by Jean-Baptiste Biot, Augustin Fresnel, and Aimé Cotton in the first half of the 19th century. Circular dichroism and optical rotation, circular birefringence are manifestations of optical activity. It is exhibited in the absorption (electromagnetic radiation), absorption bands of optical activity, optically active chirality (chemistry), chiral molecules. CD spectroscopy has a wide range of applications in many different fields. Most notably, Ultraviolet, far-UV CD is used to investigate the second ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dual Polarisation Interferometry
Dual-polarization interferometry (DPI) is an analytical technique that probes molecular layers adsorbed to the surface of a waveguide using the evanescent wave of a laser beam. It is used to measure the conformational change in proteins, or other biomolecules, as they function (referred to as the conformation activity relationship). Instrumentation DPI focuses laser light into two waveguides. One of these functions as the "sensing" waveguide having an exposed surface while the second one functions to maintain a reference beam. A two-dimensional interference pattern is formed in the far field by combining the light passing through the two waveguides. The DPI technique rotates the polarization of the laser, to alternately excite two polarization modes of the waveguides. Measurement of the interferogram for both polarizations allows both the refractive index and the thickness of the adsorbed layer to be calculated. The polarization can be switched rapidly, allowing real-time m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conformational Change
In biochemistry, a conformational change is a change in the shape of a macromolecule, often induced by environmental factors. A macromolecule is usually flexible and dynamic. Its shape can change in response to changes in its environment or other factors; each possible shape is called a conformation, and a transition between them is called a ''conformational change''. Factors that may induce such changes include temperature, pH, voltage, light in chromophores, concentration of ions, phosphorylation, or the binding of a ligand. Transitions between these states occur on a variety of length scales (tenths of Å to nm) and time scales (ns to s), and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis. Laboratory analysis Many biophysical techniques such as crystallography, NMR, electron paramagnetic resonance (EPR) using spin label techniques, circular dichroism (CD), hydrogen exchange, and FRET can be used to study macromo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
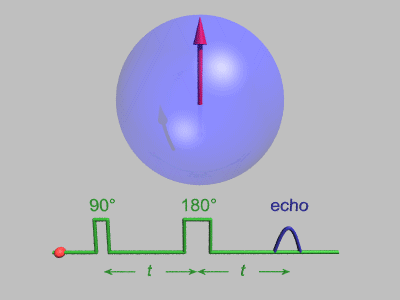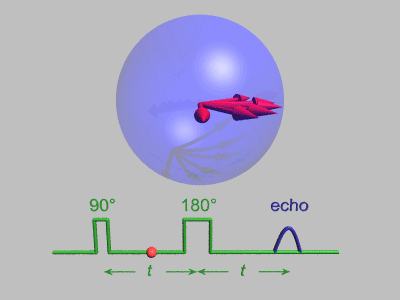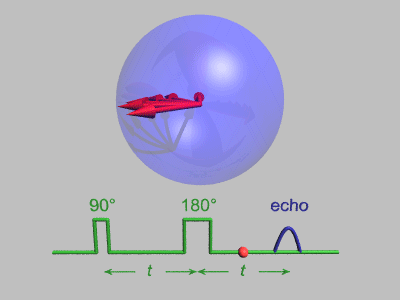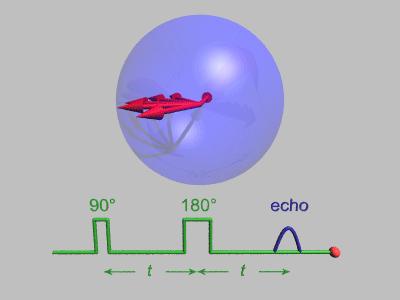
Neutron Spin Echo
Neutron spin echo spectroscopy is an inelastic neutron scattering technique invented by Ferenc Mezei in the 1970s and developed in collaboration with John Hayter. In recognition of his work and in other areas, Mezei was awarded the first Walter Haelg Prize in 1999. In magnetic resonance, a spin echo is the refocusing of spin magnetisation by a pulse of resonant electromagnetic radiation. The spin echo spectrometer possesses an extremely high energy resolution (roughly one part in 100,000). Additionally, it measures the density-density correlation (or intermediate scattering function) F(Q,t) as a function of momentum transfer Q and time. Other neutron scattering techniques measure the dynamic structure factor S(Q,ω), which can be converted to F(Q,t) by a Fourier transform, which may be difficult in practice. For weak inelastic features S(Q,ω) is better suited, however, for (slow) relaxations the natural representation is given by F(Q,t). Because of its extraordina ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dynamics
In molecular biology, proteins are generally thought to adopt unique structures determined by their amino acid sequences. However, proteins are not strictly static objects, but rather populate ensembles of (sometimes similar) conformations. Transitions between these states occur on a variety of length scales (tenths of angstroms to nm) and time scales (ns to s), and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis. The study of protein dynamics is most directly concerned with the transitions between these states, but can also involve the nature and equilibrium populations of the states themselves. These two perspectives— kinetics and thermodynamics, respectively—can be conceptually synthesized in an " energy landscape" paradigm: highly populated states and the kinetics of transitions between them can be described by the depths of energy wells and the heights of energy barriers, respectively. Local flexibility: atoms ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Small-angle Neutron Scattering
Small-angle neutron scattering (SANS) is an experimental technique that uses elastic neutron scattering at small scattering angles to investigate the structure of various substances at a mesoscopic scale of about 1–100 nm. Small angle neutron scattering is in many respects very similar to small-angle X-ray scattering (SAXS); both techniques are jointly referred to as small-angle scattering (SAS). The most important feature of the SAS method is its potential for analyzing the inner structure of disordered systems, and frequently the application of this method is a unique way to obtain direct structural information on systems with random arrangement of density inhomogeneities in such large-scales. Advantages of SANS over SAXS are its sensitivity to light elements, the possibility of isotope labelling, and the strong scattering by magnetic moments. Technique During a SANS experiment a beam of neutrons is directed at a sample, which can be an aqueous solution, a solid, a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Small-angle Scattering
Small-angle scattering (SAS) is a scattering technique based on deflection of collimated radiation away from the straight trajectory after it interacts with structures that are much larger than the wavelength of the radiation. The deflection is small (0.1-10°) hence the name ''small-angle''. SAS techniques can give information about the size, shape and orientation of structures in a sample. SAS is a powerful technique for investigating large-scale structures from 10 Å up to thousands and even several tens of thousands of angstroms. The most important feature of the SAS method is its potential for analyzing the inner structure of disordered systems, and frequently the application of this method is a unique way to obtain direct structural information on systems with random arrangement of density inhomogeneities in such large-scales. Currently, the SAS technique, with its well-developed experimental and theoretical procedures and wide range of studied objects, is a self-contain ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |