|
Mlr9623
Ku is a dimeric protein complex that binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. Ku is evolutionarily conserved from bacteria to humans. The ancestral bacterial Ku is a homodimer (two copies of the same protein bound to each other). Eukaryotic Ku is a heterodimer of two polypeptides, Ku70 (XRCC6) and Ku80 (XRCC5), so named because the molecular weight of the human Ku proteins is around 70 kDa and 80 kDa. The two Ku subunits form a basket-shaped structure that threads onto the DNA end. Once bound, Ku can slide down the DNA strand, allowing more Ku molecules to thread onto the end. In higher eukaryotes, Ku forms a complex with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the full DNA-dependent protein kinase, DNA-PK. Ku is thought to function as a molecular scaffold to which other proteins involved in NHEJ can bind, orienting the double-strand break for ligation. The Ku70 and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ku80
Ku80 is a protein that, in humans, is encoded by the ''XRCC5'' gene. Together, Ku70 and Ku80 make up the Ku heterodimer, which binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. It is also required for V(D)J recombination, which utilizes the NHEJ pathway to promote antigen diversity in the mammalian immune system. In addition to its role in NHEJ, Ku is required for telomere length maintenance and subtelomeric gene silencing. Ku was originally identified when patients with systemic lupus erythematosus were found to have high levels of autoantibodies to the protein. Nomenclature Ku80 has been referred to by several names including: * Lupus Ku autoantigen protein p80 * ATP-dependent DNA helicase 2 subunit 2 * X-ray repair complementing defective repair in Chinese hamster cells 5 * X-ray repair cross-complementing 5 (XRCC5) Epigenetic repression The protein expression level of Ku80 can be repressed by epig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Bases
Nucleobases, also known as ''nitrogenous bases'' or often simply ''bases'', are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic building blocks of nucleic acids. The ability of nucleobases to form base pairs and to stack one upon another leads directly to long-chain helical structures such as ribonucleic acid (RNA) and deoxyribonucleic acid (DNA). Five nucleobases—adenine (A), cytosine (C), guanine (G), thymine (T), and uracil (U)—are called ''primary'' or ''canonical''. They function as the fundamental units of the genetic code, with the bases A, G, C, and T being found in DNA while A, G, C, and U are found in RNA. Thymine and uracil are distinguished by merely the presence or absence of a methyl group on the fifth carbon (C5) of these heterocyclic six-membered rings. In addition, some viruses have aminoadenine (Z) instead of adenine. It differs in having an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Damage Theory Of Aging
The DNA damage theory of aging proposes that aging is a consequence of unrepaired accumulation of naturally occurring DNA damage. Damage in this context is a DNA alteration that has an abnormal structure. Although both mitochondrial and nuclear DNA damage can contribute to aging, nuclear DNA is the main subject of this analysis. Nuclear DNA damage can contribute to aging either indirectly (by increasing apoptosis or cellular senescence) or directly (by increasing cell dysfunction). Several review articles have shown that deficient DNA repair, allowing greater accumulation of DNA damage, causes premature aging; and that increased DNA repair facilitates greater longevity. Mouse models of nucleotide-excision–repair syndromes reveal a striking correlation between the degree to which specific DNA repair pathways are compromised and the severity of accelerated aging, strongly suggesting a causal relationship. Human population studies show that single-nucleotide polymorphisms in D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
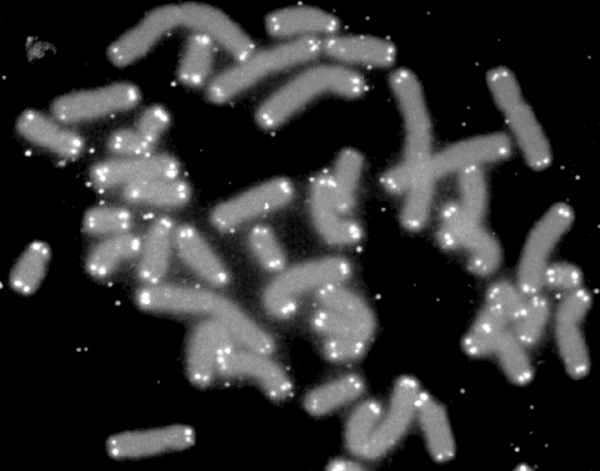
Telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes. Although there are different architectures, telomeres, in a broad sense, are a widespread genetic feature most commonly found in eukaryotes. In most, if not all species possessing them, they protect the terminal regions of chromosomal DNA from progressive degradation and ensure the integrity of linear chromosomes by preventing DNA repair systems from mistaking the very ends of the DNA strand for a double-strand break. Discovery In the early 1970s, Soviet theorist Alexei Olovnikov first recognized that chromosomes could not completely replicate their ends; this is known as the "end replication problem". Building on this, and accommodating Leonard Hayflick's idea of limited somatic cell division, Olovnikov suggested that DNA sequences are lost every time a cell replicates until the loss reaches a critical level, at which point cell division end ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosome Abnormalities
A chromosomal abnormality, chromosomal anomaly, chromosomal aberration, chromosomal mutation, or chromosomal disorder, is a missing, extra, or irregular portion of chromosomal DNA. These can occur in the form of numerical abnormalities, where there is an atypical number of chromosomes, or as structural abnormalities, where one or more individual chromosomes are altered. Chromosome mutation was formerly used in a strict sense to mean a change in a chromosomal segment, involving more than one gene. Chromosome anomalies usually occur when there is an error in cell division following meiosis or mitosis. Chromosome abnormalities may be detected or confirmed by comparing an individual's karyotype, or full set of chromosomes, to a typical karyotype for the species via genetic testing. Numerical abnormality An abnormal number of chromosomes is called aneuploidy, and occurs when an individual is either missing a chromosome from a pair (resulting in monosomy) or has more than two chromosome ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Knockout Mouse
A knockout mouse, or knock-out mouse, is a genetically modified mouse (''Mus musculus'') in which researchers have inactivated, or "knocked out", an existing gene by replacing it or disrupting it with an artificial piece of DNA. They are important animal models for studying the role of genes which have been sequenced but whose functions have not been determined. By causing a specific gene to be inactive in the mouse, and observing any differences from normal behaviour or physiology, researchers can infer its probable function. Mice are currently the laboratory animal species most closely related to humans for which the knockout technique can easily be applied. They are widely used in knockout experiments, especially those investigating genetic questions that relate to human physiology. Gene knockout in rats is much harder and has only been possible since 2003. The first recorded knockout mouse was created by Mario R. Capecchi, Martin Evans, and Oliver Smithies in 1989, for whi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Subunit
In structural biology, a protein subunit is a polypeptide chain or single protein molecule that assembles (or "''coassembles''") with others to form a protein complex. Large assemblies of proteins such as viruses often use a small number of types of protein subunits as building blocks. A subunit is often named with a Greek or Roman letter, and the numbers of this type of subunit in a protein is indicated by a subscript. For example, ATP synthase has a type of subunit called α. Three of these are present in the ATP synthase molecule, leading to the designation α3. Larger groups of subunits can also be specified, like α3β3-hexamer and c-ring. Naturally-occurring proteins that have a relatively small number of subunits are referred to as oligomeric.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminal
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an unbound amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus. Function C-terminal retention signals While the N-terminus of a protein often cont ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligases
In biochemistry, a ligase is an enzyme that can catalyze the joining (ligation) of two large molecules by forming a new chemical bond. This is typically via hydrolysis of a small pendant chemical group on one of the larger molecules or the enzyme catalyzing the linking together of two compounds, e.g., enzymes that catalyze joining of C-O, C-S, C-N, etc. In general, a ligase catalyzes the following reaction: :Ab + C → A–C + b or sometimes :Ab + cD → A–D + b + c + d + e + f where the lowercase letters can signify the small, dependent groups. Ligase can join two complementary fragments of nucleic acid and repair single stranded breaks that arise in double stranded DNA during replication. Nomenclature The common names of ligases often include the word "ligase", such as DNA ligase, an enzyme commonly used in molecular biology laboratories to join together DNA fragments. Other common names for ligases include the word "synthetase", because they are used to synthes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclease
A nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning. There are two primary classifications based on the locus of activity. Exonucleases digest nucleic acids from the ends. Endonucleases act on regions in the ''middle'' of target molecules. They are further subcategorized as deoxyribonucleases and ribonucleases. The former acts on DNA, the latter on RNA. History In the late 1960s, scientists Stuart Linn and Werner Arber isolated examples of the two types of enzymes responsible for phage growth restriction in Escherichia coli ( E. coli) bacteria. One of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerases
A polymerase is an enzyme ( EC 2.7.7.6/7/19/48/49) that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base-pairing interactions or RNA by half ladder replication. A DNA polymerase from the thermophilic bacterium, ''Thermus aquaticus'' (''Taq'') ( PDBbr>1BGX EC 2.7.7.7) is used in the polymerase chain reaction, an important technique of molecular biology. A polymerase may be template dependent or template independent. Poly-A-polymerase is an example of template independent polymerase. Terminal deoxynucleotidyl transferase also known to have template independent and template dependent activities. Types By function *DNA polymerase (DNA-directed DNA polymerase, DdDP) **Family A: DNA polymerase I; Pol γ, θ, ν **Family B: DNA polymerase II; Pol α, δ, ε, ζ **Family C: DNA polymerase III holoenzyme **Family X: Pol β, λ, μ * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structurally
A structure is an arrangement and organization of interrelated elements in a material object or system, or the object or system so organized. Material structures include man-made objects such as buildings and machines and natural objects such as biological organisms, minerals and chemicals. Abstract structures include data structures in computer science and musical form. Types of structure include a hierarchy (a cascade of one-to-many relationships), a network featuring many-to-many links, or a lattice featuring connections between components that are neighbors in space. Load-bearing Buildings, aircraft, skeletons, anthills, beaver dams, bridges and salt domes are all examples of load-bearing structures. The results of construction are divided into buildings and non-building structures, and make up the infrastructure of a human society. Built structures are broadly divided by their varying design approaches and standards, into categories including building structures, archi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |