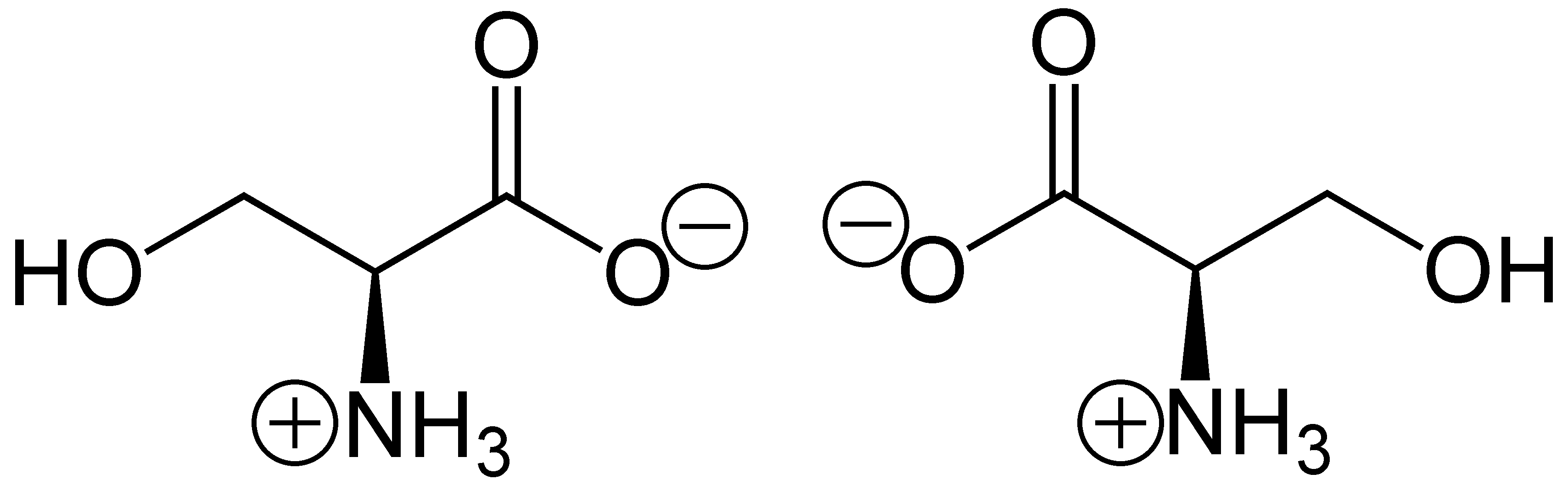|
Metallopeptidase
A metalloproteinase, or metalloprotease, is any protease enzyme whose catalytic mechanism involves a metal. An example is ADAM12 which plays a significant role in the fusion of muscle cells during embryo development, in a process known as myogenesis. Most metalloproteases require zinc, but some use cobalt. The metal ion is coordinated to the protein via three ligands. The ligands coordinating the metal ion can vary with histidine, glutamate, aspartate, lysine, and arginine. The fourth coordination position is taken up by a labile water molecule. Treatment with chelating agents such as EDTA leads to complete inactivation. EDTA is a metal chelator that removes zinc, which is essential for activity. They are also inhibited by the chelator orthophenanthroline. Classification There are two subgroups of metalloproteinases: * Exopeptidases, metalloexopeptidases ( EC number: 3.4.17). * Endopeptidases, metalloendopeptidases (3.4.24). Well known metalloendopeptidases include ADAM pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Hierarchy of proteases Based on catalytic residue Proteases can be classified into seven broad groups: * Serine protease ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EDTA
Ethylenediaminetetraacetic acid (EDTA) is an aminopolycarboxylic acid with the formula H2N(CH2CO2H)2sub>2. This white, water-soluble solid is widely used to bind to iron (Fe2+/Fe3+) and calcium ions (Ca2+), forming water-soluble complexes even at neutral pH. It is thus used to dissolve Fe- and Ca-containing scale as well as to deliver iron ions under conditions where its oxides are insoluble. EDTA is available as several salts, notably disodium EDTA, sodium calcium edetate, and tetrasodium EDTA, but these all function similarly. Uses Textile industry In industry, EDTA is mainly used to sequester (bind or confine) metal ions in aqueous solution. In the textile industry, it prevents metal ion impurities from modifying colours of dyed products. In the pulp and paper industry, EDTA inhibits the ability of metal ions, especially Mn2+, from catalysing the disproportionation of hydrogen peroxide, which is used in chlorine-free bleaching. In a similar manner, EDTA is added ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Threonine
Threonine (symbol Thr or T) is an amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), a carboxyl group (which is in the deprotonated −COO− form under biological conditions), and a side chain containing a hydroxyl group, making it a polar, uncharged amino acid. It is essential in humans, meaning the body cannot synthesize it: it must be obtained from the diet. Threonine is synthesized from aspartate in bacteria such as ''E. coli''. It is encoded by all the codons starting AC (ACU, ACC, ACA, and ACG). Threonine sidechains are often hydrogen bonded; the most common small motifs formed are based on interactions with serine: ST turns, ST motifs (often at the beginning of alpha helices) and ST staples (usually at the middle of alpha helices). Modifications The threonine residue is susceptible to numerous posttranslational modifications. The hydroxyl side-chain can unde ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L-stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure was estab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cysteine
Cysteine (symbol Cys or C; ) is a semiessential proteinogenic amino acid with the formula . The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile. When present as a deprotonated catalytic residue, sometimes the symbol Cyz is used. The deprotonated form can generally be described by the symbol Cym as well. The thiol is susceptible to oxidation to give the disulfide derivative cystine, which serves an important structural role in many proteins. In this case, the symbol Cyx is sometimes used. When used as a food additive, it has the E number E920. Cysteine is encoded by the codons UGU and UGC. The sulfur-containing amino acids cysteine and methionine are more easily oxidized than the other amino acids. Structure Like other amino acids (not as a residue of a protein), cysteine exists as a zwitterion. Cysteine has chirality in the older / notation based on homology to - and -glyceraldehyde. In the newer ''R''/''S'' system of designating chi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MEROPS Database
MEROPS is an online database for peptidases (also known as proteases, proteinases and proteolytic enzymes) and their inhibitors. The classification scheme for peptidases was published by Rawlings & Barrett in 1993, and that for protein inhibitors by Rawlings ''et al.'' in 2004.Rawlings, N.D., Tolle, D.P. & Barrett, A.J. (2004) "Evolutionary families of peptidase inhibitors." ''Biochem J'' 378, 705-716. The most recent version, MEROPS 12.3, was released in September 2020. Overview The classification is based on similarities at the tertiary and primary structural levels. Comparisons are restricted to that part of the sequence directly involved in the reaction, which in the case of a peptidase must include the active site, and for a protein inhibitor the reactive site. The classification is hierarchical: sequences are assembled into families, and families are assembled into clans. Each peptidase, family, and clan has a unique identifier. Classification Family The families of pe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Matrix Metalloproteinase
Matrix metalloproteinases (MMPs), also known as matrix metallopeptidases or matrixins, are metalloproteinases that are calcium-dependent zinc-containing endopeptidases; other family members are adamalysins, serralysins, and astacins. The MMPs belong to a larger family of proteases known as the metzincin superfamily. Collectively, these enzymes are capable of degrading all kinds of extracellular matrix proteins, but also can process a number of bioactive molecules. They are known to be involved in the cleavage of cell surface receptors, the release of apoptotic ligands (such as the FAS ligand), and chemokine/cytokine inactivation. MMPs are also thought to play a major role in cell behaviors such as cell proliferation, migration (adhesion/dispersion), differentiation, angiogenesis, apoptosis, and host defense. They were first described in vertebrates (1962), including humans, but have since been found in invertebrates and plants. They are distinguished from other endopeptida ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ADAM Protein
ADAMs (short for a disintegrin and metalloproteinase) are a family of single-pass transmembrane and secreted metalloendopeptidases. All ADAMs are characterized by a particular domain organization featuring a pro-domain, a metalloprotease, a disintegrin, a cysteine-rich, an epidermal-growth factor like and a transmembrane domain, as well as a C-terminal cytoplasmic tail. Nonetheless, not all human ADAMs have a functional protease domain, which indicates that their biological function mainly depends on protein–protein interactions. Those ADAMs which are active proteases are classified as sheddases because they cut off or shed extracellular portions of transmembrane proteins. For example, ADAM10 can cut off part of the HER2 receptor, thereby activating it. ADAM genes are found in animals, choanoflagellates, fungi and some groups of green algae. Most green algae and all land plants likely lost ADAM proteins. ADAMs are categorized under the enzyme group, and in the MEROPS peptidas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metalloendopeptidase
A metalloendopeptidase is an enzyme that functions as a metalloproteinase endopeptidase Endopeptidase or endoproteinase are proteolytic peptidases that break peptide bonds of nonterminal amino acids (i.e. within the molecule), in contrast to exopeptidases, which break peptide bonds from end-pieces of terminal amino acids. For this re .... References External links * EC 3.4.24 {{enzyme-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endopeptidase
Endopeptidase or endoproteinase are proteolytic peptidases that break peptide bonds of nonterminal amino acids (i.e. within the molecule), in contrast to exopeptidases, which break peptide bonds from end-pieces of terminal amino acids. For this reason, endopeptidases cannot break down peptides into monomers, while exopeptidases can break down proteins into monomers. A particular case of endopeptidase is the oligopeptidase, whose substrates are oligopeptides instead of proteins. They are usually very specific for certain amino acids. Examples of endopeptidases include: * Trypsin - cuts after Arg or Lys, unless followed by Pro. Very strict. Works best at pH 8. * Chymotrypsin - cuts after Phe, Trp, or Tyr, unless followed by Pro. Cuts more slowly after His, Met or Leu. Works best at pH 8. * Elastase - cuts after Ala, Gly, Ser, or Val, unless followed by Pro. * Thermolysin - cuts ''before'' Ile, Met, Phe, Trp, Tyr, or Val, unless ''preceded'' by Pro. Sometimes cuts after Ala, Asp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme Commission Number
The Enzyme Commission number (EC number) is a numerical classification scheme for enzymes, based on the chemical reactions they catalyze. As a system of enzyme nomenclature, every EC number is associated with a recommended name for the corresponding enzyme-catalyzed reaction. EC numbers do not specify enzymes but enzyme-catalyzed reactions. If different enzymes (for instance from different organisms) catalyze the same reaction, then they receive the same EC number. Furthermore, through convergent evolution, completely different protein folds can catalyze an identical reaction (these are sometimes called non-homologous isofunctional enzymes) and therefore would be assigned the same EC number. By contrast, UniProt identifiers uniquely specify a protein by its amino acid sequence. Format of number Every enzyme code consists of the letters "EC" followed by four numbers separated by periods. Those numbers represent a progressively finer classification of the enzyme. Preliminary EC ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |