|
Local Elevation
Local elevation is a technique used in computational chemistry or physics, mainly in the field of molecular simulation (including molecular dynamics (MD) and Monte Carlo (MC) simulations). It was developed in 1994 by Huber, Torda and van Gunsteren to enhance the searching of conformational space in molecular dynamics simulations and is available in the GROMOS software for molecular dynamics simulation (since GROMOS96). The method was, together with the conformational flooding method, the first to introduce memory dependence into molecular simulations. Many recent methods build on the principles of the local elevation technique, including the Engkvist-Karlström, adaptive biasing force, Wang–Landau, metadynamics, adaptively biased molecular dynamics, adaptive reaction coordinate forces, and local elevation umbrella sampling methods. The basic principle of the method is to add a memory-dependent potential energy term in the simulation so as to prevent the simulation to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Computational Chemistry
Computational chemistry is a branch of chemistry that uses computer simulation to assist in solving chemical problems. It uses methods of theoretical chemistry, incorporated into computer programs, to calculate the structures and properties of molecules, groups of molecules, and solids. It is essential because, apart from relatively recent results concerning the hydrogen molecular ion (dihydrogen cation, see references therein for more details), the quantum many-body problem cannot be solved analytically, much less in closed form. While computational results normally complement the information obtained by chemical experiments, it can in some cases predict hitherto unobserved chemical phenomena. It is widely used in the design of new drugs and materials. Examples of such properties are structure (i.e., the expected positions of the constituent atoms), absolute and relative (interaction) energies, electronic charge density distributions, dipoles and higher multipole moments, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Computational Physics
Computational physics is the study and implementation of numerical analysis to solve problems in physics for which a quantitative theory already exists. Historically, computational physics was the first application of modern computers in science, and is now a subset of computational science. It is sometimes regarded as a subdiscipline (or offshoot) of theoretical physics, but others consider it an intermediate branch between theoretical and experimental physics - an area of study which supplements both theory and experiment. Overview In physics, different theories based on mathematical models provide very precise predictions on how systems behave. Unfortunately, it is often the case that solving the mathematical model for a particular system in order to produce a useful prediction is not feasible. This can occur, for instance, when the solution does not have a closed-form expression, or is too complicated. In such cases, numerical approximations are required. Computational phy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Dynamics
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics. Because molecular systems typically consist of a vast number of particles, it is impossible to determine the properties of such complex systems analytically; MD simulation circumvents this problem by using numerical methods. However, long MD simulations are mathematically ill-conditioned, generating cumulative erro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
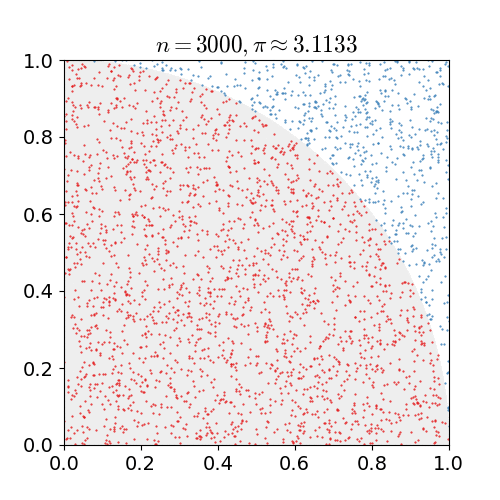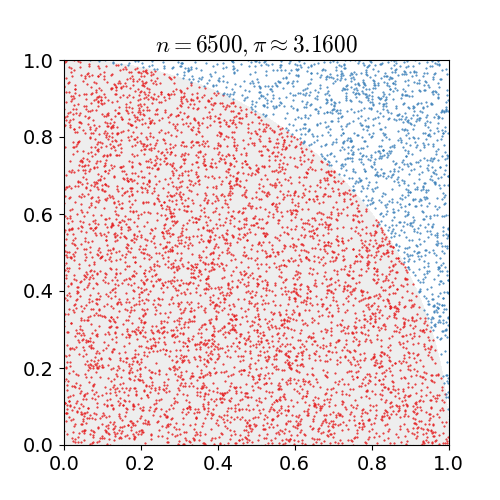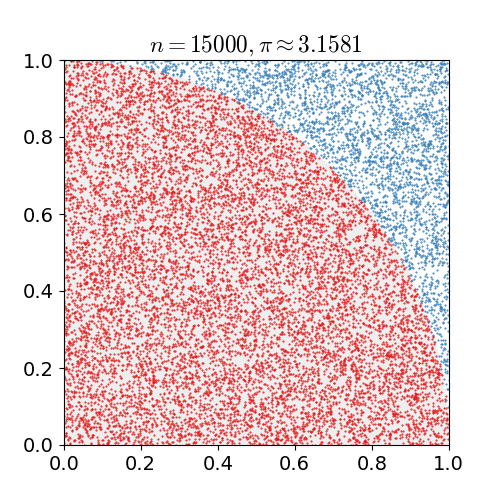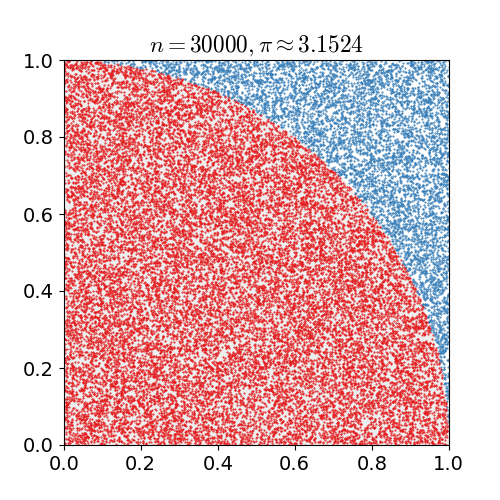
Monte Carlo Method
Monte Carlo methods, or Monte Carlo experiments, are a broad class of computational algorithms that rely on repeated random sampling to obtain numerical results. The underlying concept is to use randomness to solve problems that might be deterministic in principle. They are often used in physical and mathematical problems and are most useful when it is difficult or impossible to use other approaches. Monte Carlo methods are mainly used in three problem classes: optimization, numerical integration, and generating draws from a probability distribution. In physics-related problems, Monte Carlo methods are useful for simulating systems with many coupled degrees of freedom, such as fluids, disordered materials, strongly coupled solids, and cellular structures (see cellular Potts model, interacting particle systems, McKean–Vlasov processes, kinetic models of gases). Other examples include modeling phenomena with significant uncertainty in inputs such as the calculation of ris ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GROMOS
GROningen MOlecular Simulation (GROMOS) is the name of a force field for molecular dynamics simulation, and a related computer software package. Both are developed at the University of Groningen, and at the Computer-Aided Chemistry Group at the Laboratory for Physical Chemistry at the Swiss Federal Institute of Technology (ETH Zurich). At Groningen, Herman Berendsen was involved in its development. The united atom force field was optimized with respect to the condensed phase properties of alkanes. Versions GROMOS87 Aliphatic and aromatic hydrogen atoms were included implicitly by representing the carbon atom and attached hydrogen atoms as one group centered on the carbon atom, a united atom force field. The van der Waals force parameters were derived from calculations of the crystal structures of hydrocarbons, and on amino acids using short (0.8 nm) nonbonded cutoff radii. GROMOS96 In 1996, a substantial rewrite of the software package was released. The force fiel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Wang And Landau Algorithm
The Wang and Landau algorithm, proposed by Fugao Wang and David P. Landau, is a Monte Carlo method designed to estimate the density of states of a system. The method performs a non-Markovian random walk to build the density of states by quickly visiting all the available energy spectrum. The Wang and Landau algorithm is an important method to obtain the density of states required to perform a multicanonical simulation. The Wang–Landau algorithm can be applied to any system which is characterized by a cost (or energy) function. For instance, it has been applied to the solution of numerical integrals and the folding of proteins. The Wang–Landau sampling is related to the metadynamics algorithm.Christoph Junghans, Danny Perez, and Thomas Vogel. "Molecular Dynamics in the Multicanonical Ensemble: Equivalence of Wang–Landau Sampling, Statistical Temperature Molecular Dynamics, and Metadynamics." Journal of Chemical Theory and Computation 10.5 (2014): 1843-1847. doibr>10.1021/c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metadynamics
Metadynamics (MTD; also abbreviated as METAD or MetaD) is a computer simulation method in computational physics, chemistry and biology. It is used to estimate the free energy and other state functions of a system, where ergodicity is hindered by the form of the system's energy landscape. It was first suggested by Alessandro Laio and Michele Parrinello in 2002 and is usually applied within molecular dynamics simulations. MTD closely resembles a number of recent methods such as adaptively biased molecular dynamics, adaptive reaction coordinate forces and local elevation umbrella sampling. More recently, both the original and well-tempered metadynamics were derived in the context of importance sampling and shown to be a special case of the adaptive biasing potential setting. MTD is related to the Wang–Landau sampling. Introduction The technique builds on a large number of related methods including (in a chronological order) the deflation, tunneling, tabu search, local eleva ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tabu Search
Tabu search is a metaheuristic search method employing local search methods used for mathematical optimization. It was created by Fred W. Glover in 1986 and formalized in 1989. Local (neighborhood) searches take a potential solution to a problem and check its immediate neighbors (that is, solutions that are similar except for very few minor details) in the hope of finding an improved solution. Local search methods have a tendency to become stuck in suboptimal regions or on plateaus where many solutions are equally fit. Tabu search enhances the performance of local search by relaxing its basic rule. First, at each step ''worsening'' moves can be accepted if no improving move is available (like when the search is stuck at a strict local minimum). In addition, ''prohibitions'' (henceforth the term ''tabu'') are introduced to discourage the search from coming back to previously-visited solutions. The implementation of tabu search uses memory structures that describe the visited ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
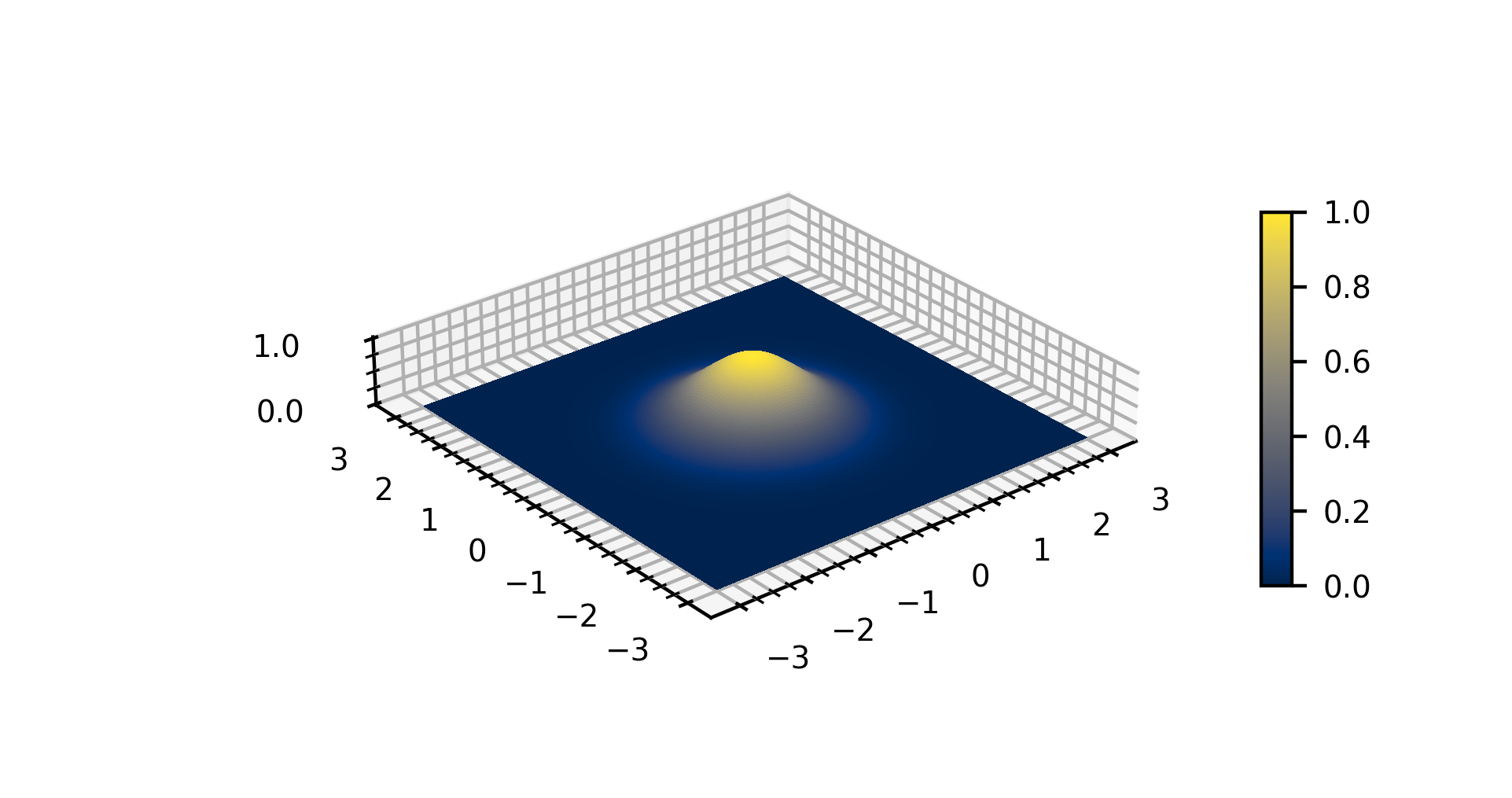
Gaussian Function
In mathematics, a Gaussian function, often simply referred to as a Gaussian, is a function of the base form f(x) = \exp (-x^2) and with parametric extension f(x) = a \exp\left( -\frac \right) for arbitrary real constants , and non-zero . It is named after the mathematician Carl Friedrich Gauss. The graph of a Gaussian is a characteristic symmetric " bell curve" shape. The parameter is the height of the curve's peak, is the position of the center of the peak, and (the standard deviation, sometimes called the Gaussian RMS width) controls the width of the "bell". Gaussian functions are often used to represent the probability density function of a normally distributed random variable with expected value and variance . In this case, the Gaussian is of the form g(x) = \frac \exp\left( -\frac \frac \right). Gaussian functions are widely used in statistics to describe the normal distributions, in signal processing to define Gaussian filters, in image processing where two-dimen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polynomial
In mathematics, a polynomial is an expression consisting of indeterminates (also called variables) and coefficients, that involves only the operations of addition, subtraction, multiplication, and positive-integer powers of variables. An example of a polynomial of a single indeterminate is . An example with three indeterminates is . Polynomials appear in many areas of mathematics and science. For example, they are used to form polynomial equations, which encode a wide range of problems, from elementary word problems to complicated scientific problems; they are used to define polynomial functions, which appear in settings ranging from basic chemistry and physics to economics and social science; they are used in calculus and numerical analysis to approximate other functions. In advanced mathematics, polynomials are used to construct polynomial rings and algebraic varieties, which are central concepts in algebra and algebraic geometry. Etymology The word ''polynomial'' joins tw ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Umbrella Sampling
Umbrella sampling is a technique in computational physics and chemistry, used to improve sampling of a system (or different systems) where ergodicity is hindered by the form of the system's energy landscape. It was first suggested by Torrie and Valleau in 1977. It is a particular physical application of the more general importance sampling in statistics. Systems in which an energy barrier separates two regions of configuration space may suffer from poor sampling. In Metropolis Monte Carlo runs, the low probability of overcoming the potential barrier can leave inaccessible configurations poorly sampled—or even entirely unsampled—by the simulation. An easily visualised example occurs with a solid at its melting point: considering the state of the system with an order parameter ''Q'', both liquid (low ''Q'') and solid (high ''Q'') phases are low in energy, but are separated by a free energy barrier at intermediate values of ''Q''. This prevents the simulation from adequatel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Dynamics
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics. Because molecular systems typically consist of a vast number of particles, it is impossible to determine the properties of such complex systems analytically; MD simulation circumvents this problem by using numerical methods. However, long MD simulations are mathematically ill-conditioned, generating cumulative erro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |