|
GROMOS
GROningen MOlecular Simulation (GROMOS) is the name of a force field for molecular dynamics simulation, and a related computer software package. Both are developed at the University of Groningen, and at the Computer-Aided Chemistry Group at the Laboratory for Physical Chemistry at the Swiss Federal Institute of Technology (ETH Zurich). At Groningen, Herman Berendsen was involved in its development. The united atom force field was optimized with respect to the condensed phase properties of alkanes. Versions GROMOS87 Aliphatic and aromatic hydrogen atoms were included implicitly by representing the carbon atom and attached hydrogen atoms as one group centered on the carbon atom, a united atom force field. The van der Waals force parameters were derived from calculations of the crystal structures of hydrocarbons, and on amino acids using short (0.8 nm) nonbonded cutoff radii. GROMOS96 In 1996, a substantial rewrite of the software package was released. The force fiel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Force Field (chemistry)
In the context of chemistry and molecular modelling, a force field is a computational method that is used to estimate the forces between atoms within molecules and also between molecules. More precisely, the force field refers to the functional form and parameter sets used to calculate the potential energy of a system of atoms or coarse-grained particles in molecular mechanics, molecular dynamics, or Monte Carlo simulations. The parameters for a chosen energy function may be derived from experiments in physics and chemistry, calculations in quantum mechanics, or both. Force fields are interatomic potentials and utilize the same concept as force fields in classical physics, with the difference that the force field parameters in chemistry describe the energy landscape, from which the acting forces on every particle are derived as a gradient of the potential energy with respect to the particle coordinates. ''All-atom'' force fields provide parameters for every type of atom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparison Of Software For Molecular Mechanics Modeling
This is a list of computer programs that are predominantly used for molecular mechanics calculations. See also * Car–Parrinello molecular dynamics * Comparison of force-field implementations *Comparison of nucleic acid simulation software * List of molecular graphics systems *List of protein structure prediction software *List of quantum chemistry and solid-state physics software *List of software for Monte Carlo molecular modeling * List of software for nanostructures modeling * Molecular design software *Molecular dynamics *Molecular modeling on GPUs Molecular modeling on GPU is the technique of using a graphics processing unit (GPU) for molecular simulations. In 2007, NVIDIA introduced video cards that could be used not only to show graphics but also for scientific calculations. These cards ... * Molecule editor Notes and references External linksSINCRIS [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Herman Berendsen
Herman Johan Christiaan Berendsen (22 September 1934 – 7 October 2019) was a Dutch chemist. He was a professor of physical chemistry at the University of Groningen from 1967 to 1999. Career Berendsen was born on 22 September 1934 in Apeldoorn. In 1962 he obtained his PhD ''cum laude'' from the University of Groningen, with a dissertation titled: "An NMR study of collagen hydration". In March 1963 Berendsen was appointed lector of physical chemistry at his ''alma mater'', and four years later became full professor. He retired in 1999. At Groningen Berendsen was group leader of molecular dynamics. In 1976 Berendsen and Wilfred van Gunsteren together with the 2013 Nobel Prize in Chemistry winners Martin Karplus and Michael Levitt were involved in the start of theories on molecular calculating. Berendsen was elected a member of the Royal Netherlands Academy of Arts and Sciences in 1979. In 2013 he was awarded the Berni J. Alder Prize by the Centre européen de calcul atomique ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
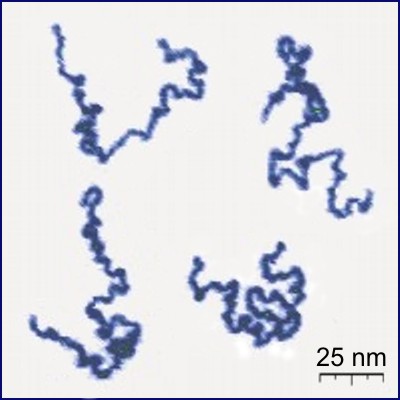
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C++ Software
C, or c, is the third letter in the Latin alphabet, used in the modern English alphabet, the alphabets of other western European languages and others worldwide. Its name in English is ''cee'' (pronounced ), plural ''cees''. History "C" comes from the same letter as "G". The Semites named it gimel. The sign is possibly adapted from an Egyptian hieroglyph for a staff sling, which may have been the meaning of the name ''gimel''. Another possibility is that it depicted a camel, the Semitic name for which was ''gamal''. Barry B. Powell, a specialist in the history of writing, states "It is hard to imagine how gimel = "camel" can be derived from the picture of a camel (it may show his hump, or his head and neck!)". In the Etruscan language, plosive consonants had no contrastive voicing, so the Greek ' Γ' (Gamma) was adopted into the Etruscan alphabet to represent . Already in the Western Greek alphabet, Gamma first took a '' form in Early Etruscan, then '' in Classical Et ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparison Of Force Field Implementations
This is a table of notable computer programs implementing molecular mechanics force fields. See also *Force field (chemistry) * List of software for Monte Carlo molecular modeling *Molecular mechanics * Molecular design software *Molecule editor *Comparison of software for molecular mechanics modeling This is a list of computer programs that are predominantly used for molecular mechanics calculations. See also * Car–Parrinello molecular dynamics * Comparison of force-field implementations *Comparison of nucleic acid simulation software ... * Molecular modeling on GPU References {{Reflist Force fields Molecular modelling Software comparisons ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ascalaph Designer
Ascalaph Designer is a computer program for general purpose molecular modelling for molecular design and simulations. It provides a graphical environment for the common programs of quantum and classical molecular modelling ORCA, NWChem, Firefly, CP2K and MDynaMix . The molecular mechanics calculations cover model building, energy optimizations and molecular dynamics. Firefly (formerly named PC GAMESS) covers a wide range of quantum chemistry methods. Ascalaph Designer is free and open-source software, released under the GNU General Public License, version 2 (GPLv2). Key features Uses See also * List of software for molecular mechanics modeling * Molecular design software * Molecule editor * Abalone Abalone ( or ; via Spanish , from Rumsen ''aulón'') is a common name for any of a group of small to very large marine gastropod molluscs in the family Haliotidae. Other common names are ear shells, sea ears, and, rarely, muttonfish or mutto ... References Ext ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymer
A polymer (; Greek '' poly-'', "many" + ''-mer'', "part") is a substance or material consisting of very large molecules called macromolecules, composed of many repeating subunits. Due to their broad spectrum of properties, both synthetic and natural polymers play essential and ubiquitous roles in everyday life. Polymers range from familiar synthetic plastics such as polystyrene to natural biopolymers such as DNA and proteins that are fundamental to biological structure and function. Polymers, both natural and synthetic, are created via polymerization of many small molecules, known as monomers. Their consequently large molecular mass, relative to small molecule compounds, produces unique physical properties including toughness, high elasticity, viscoelasticity, and a tendency to form amorphous and semicrystalline structures rather than crystals. The term "polymer" derives from the Greek word πολύς (''polus'', meaning "many, much") and μέρος (''meros'' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Micelle
A micelle () or micella () (plural micelles or micellae, respectively) is an aggregate (or supramolecular assembly) of surfactant amphipathic lipid molecules dispersed in a liquid, forming a colloidal suspension (also known as associated colloidal system). A typical micelle in water forms an aggregate with the hydrophilic "head" regions in contact with surrounding solvent, sequestering the hydrophobic single-tail regions in the micelle centre. This phase is caused by the packing behavior of single-tail lipids in a bilayer. The difficulty filling all the volume of the interior of a bilayer, while accommodating the area per head group forced on the molecule by the hydration of the lipid head group, leads to the formation of the micelle. This type of micelle is known as a normal-phase micelle (oil-in-water micelle). Inverse micelles have the head groups at the centre with the tails extending out (water-in-oil micelle). Micelles are approximately spherical in shape. Other phas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Membrane
A membrane is a selective barrier; it allows some things to pass through but stops others. Such things may be molecules, ions, or other small particles. Membranes can be generally classified into synthetic membranes and biological membranes. Biological membranes include cell membranes (outer coverings of cells or organelles that allow passage of certain constituents); nuclear membranes, which cover a cell nucleus; and tissue membranes, such as mucosae and serosae. Synthetic membranes are made by humans for use in laboratories and industry (such as chemical plants). This concept of a membrane has been known since the eighteenth century but was used little outside of the laboratory until the end of World War II. Drinking water supplies in Europe had been compromised by the war and membrane filters were used to test for water safety. However, due to the lack of reliability, slow operation, reduced selectivity and elevated costs, membranes were not widely exploited. The first ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |