|
Lysine Carboxypeptidase
Lysine carboxypeptidase ( EC 3.4.17.3) is an enzyme. This enzyme catalyses the following chemical reaction: : Release of a C-terminal basic amino acid (lysine or arginine), preferentially lysine. This is a zinc-activated enzyme found in plasma. It inactivates proteins such as bradykinin and anaphylatoxins in the blood in order to prevent toxic buildup. Nomenclature Lysine carboxypeptidase is also known as: * carboxypeptidase N * arginine carboxypeptidase * kininase I * anaphylatoxin inactivator * plasma carboxypeptidase B * creatine kinase conversion factor * bradykinase * kininase Ia * hippuryllysine hydrolase * bradykinin-decomposing enzyme * protaminase * CPase N * creatinine kinase convertase * peptidyl-L-lysine(-L-arginine) hydrolase * CPN Classification All enzymes are assigned an Enzyme Commission number based on the chemical reaction they catalyze. An EC number functions to clear up any confusion that arises due to the fact that many enzymes have s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme Commission Number
The Enzyme Commission number (EC number) is a numerical classification scheme for enzymes, based on the chemical reactions they catalyze. As a system of enzyme nomenclature, every EC number is associated with a recommended name for the corresponding enzyme-catalyzed reaction. EC numbers do not specify enzymes but enzyme-catalyzed reactions. If different enzymes (for instance from different organisms) catalyze the same reaction, then they receive the same EC number. Furthermore, through convergent evolution, completely different protein folds can catalyze an identical reaction (these are sometimes called non-homologous isofunctional enzymes) and therefore would be assigned the same EC number. By contrast, UniProt identifiers uniquely specify a protein by its amino acid sequence. Format of number Every enzyme code consists of the letters "EC" followed by four numbers separated by periods. Those numbers represent a progressively finer classification of the enzyme. Preliminary EC ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
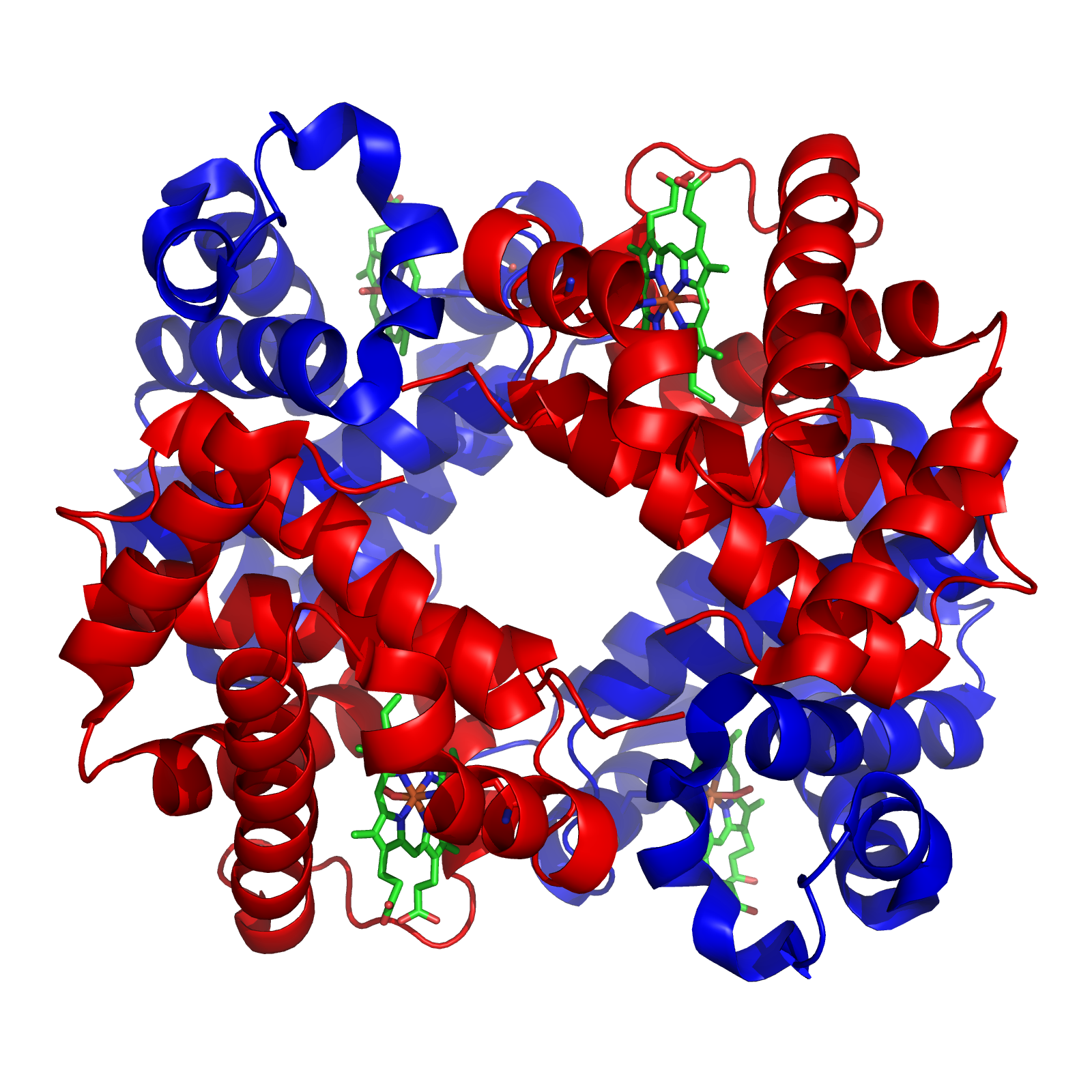
Tetramer
A tetramer () (''tetra-'', "four" + '' -mer'', "parts") is an oligomer formed from four monomers or subunits. The associated property is called ''tetramery''. An example from inorganic chemistry is titanium methoxide with the empirical formula Ti(OCH3)4, which is tetrameric in solid state and has the molecular formula Ti4(OCH3)16. An example from organic chemistry is kobophenol A, a substance that is formed by combining four molecules of resveratrol. In biochemistry, it similarly refers to a biomolecule formed of four units, that are the same (homotetramer), i.e. as in Concanavalin A or different (heterotetramer), i.e. as in hemoglobin. Hemoglobin has 4 similar sub-units while immunoglobulins have 2 very different sub-units. The different sub-units may have each their own activity, such as binding biotin in avidin tetramers, or have a common biological property, such as the allosteric binding of oxygen in hemoglobin. See also * Cluster chemistry; atomic and molecular clusters ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrophobe
In chemistry, hydrophobicity is the physical property of a molecule that is seemingly repelled from a mass of water (known as a hydrophobe). In contrast, hydrophiles are attracted to water. Hydrophobic molecules tend to be nonpolar and, thus, prefer other neutral molecules and nonpolar solvents. Because water molecules are polar, hydrophobes do not dissolve well among them. Hydrophobic molecules in water often cluster together, forming micelles. Water on hydrophobic surfaces will exhibit a high contact angle. Examples of hydrophobic molecules include the alkanes, oils, fats, and greasy substances in general. Hydrophobic materials are used for oil removal from water, the management of oil spills, and chemical separation processes to remove non-polar substances from polar compounds. Hydrophobic is often used interchangeably with lipophilic, "fat-loving". However, the two terms are not synonymous. While hydrophobic substances are usually lipophilic, there are exceptions, suc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antiparallel (biochemistry)
In biochemistry, two biopolymers are antiparallel if they run parallel to each other but with opposite directionality (alignments). An example is the two complementary strands of a DNA double helix, which run in opposite directions alongside each other. Nucleic acids Nucleic acid molecules have a phosphoryl (5') end and a hydroxyl (3') end. This notation follows from organic chemistry nomenclature, and can be used to define the movement of enzymes such as DNA polymerases relative to the DNA strand in a non-arbitrary manner. G-quadruplexes G-quadruplexes, also known as G4 DNA are secondary structures found in nucleic acids that are rich in guanine. These structures are normally located at the telomeres (the ends of the chromosomes). The G-quadruplex can either be parallel or antiparallel depending on the loop configuration, which is a component of the structure. If all the DNA strands run in the same direction, it is termed to be a parallel quadruplex, and is known as a str ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disulfide
In biochemistry, a disulfide (or disulphide in British English) refers to a functional group with the structure . The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. ''Persulfide'' usually refers to compounds. In inorganic chemistry disulfide usually refers to the corresponding anion (−S−S−). Organic disulfides Symmetrical disulfides are compounds of the formula . Most disulfides encountered in organo sulfur chemistry are symmetrical disulfides. Unsymmetrical disulfides (also called heterodisulfides) are compounds of the formula . They are less common in organic chemistry, but most disulfides in nature are unsymmetrical. Properties The disulfide bonds are strong, with a typical bond dissociation energy of 60 kcal/mol (251&nbs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Turn
β turns (also β-bends, tight turns, reverse turns, Venkatachalam turns) are the most common form of turns—a type of non-regular secondary structure in proteins that cause a change in direction of the polypeptide chain. They are very common motifs in proteins and polypeptides. Each consists of four amino acid residues (labelled ''i'', ''i+1'', ''i+2'' and ''i+3''). They can be defined in two ways: # By the possession of an intra-main-chain hydrogen bond between the CO of residue ''i'' and the NH of residue ''i+3''; # By having a distance of less than 7Å between the Cα atoms of residues ''i'' and ''i+3''. The hydrogen bond criterion is the one most appropriate for everyday use, partly because it gives rise to four distinct categories; the distance criterion gives rise to the same four categories but yields additional turn types. Definition Hydrogen bond criterion The hydrogen bond criterion for beta turns, applied to polypeptides whose amino acids are linked by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined versi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ESyPred3D
ESyPred3D is an automated homology modeling program. Alignments are obtained by combining, weighting and screening the results of several multiple alignment programs. The final three-dimensional structure is built using the modeling package MODELLER. Method To perform homology modeling, the ESyPred3D program first searches for a template (a similar sequence of known structure), before aligning the query and template sequences. EsyPred3D then build the 3D models using the alignment and the template structure, before assessing the final 3D model. The query and the template sequences are aligned using a consensus alignment method. Different multiple sequence alignments are built using different alignment programs on two sets of sequences including the query and the template sequence. The consensus method uses a neural network to find the best aligned residues and analyzing all possible combinations using a dead end elimination algorithm. The final 3D model is built from the target ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Leucine-rich Repeat
A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand- turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues. Leucine-rich repeats are frequently involved in the formation of protein–protein interactions. Examples Leucine-rich repeat motifs have been identified in a large numbe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cysteine
Cysteine (symbol Cys or C; ) is a semiessential proteinogenic amino acid with the formula . The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile. When present as a deprotonated catalytic residue, sometimes the symbol Cyz is used. The deprotonated form can generally be described by the symbol Cym as well. The thiol is susceptible to oxidation to give the disulfide derivative cystine, which serves an important structural role in many proteins. In this case, the symbol Cyx is sometimes used. When used as a food additive, it has the E number E920. Cysteine is encoded by the codons UGU and UGC. The sulfur-containing amino acids cysteine and methionine are more easily oxidized than the other amino acids. Structure Like other amino acids (not as a residue of a protein), cysteine exists as a zwitterion. Cysteine has chirality in the older / notation based on homology to - and -glyceraldehyde. In the newer ''R''/''S'' system of designating chi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |

_structure.png)



