|
Lung Microbiota
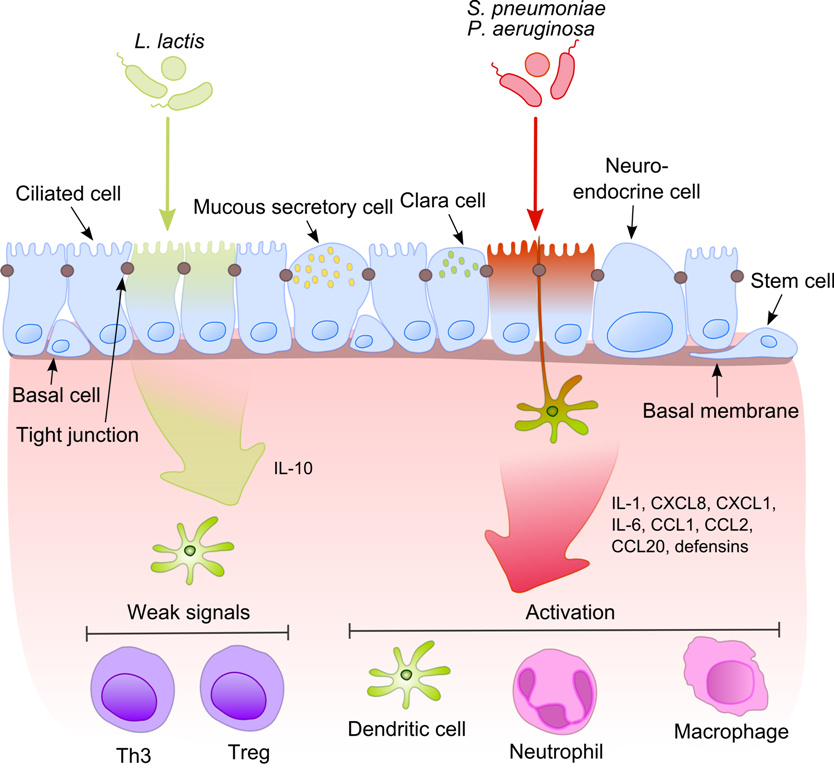
The lung microbiota, is the pulmonary microbial community consisting of a complex variety of microorganisms found in the lower respiratory tract particularly on the mucous layer and the epithelial surfaces. These microorganisms include bacteria, fungi, viruses and bacteriophages. The bacterial part of the microbiota has been more closely studied. It consists of a core of nine genera: ''Prevotella'', ''Sphingomonas'', ''Pseudomonas'', ''Acinetobacter'', ''Fusobacterium'', ''Megasphaera'', ''Veillonella'', '' Staphylococcus'', and ''Streptococcus''. They are aerobes as well as anaerobes and aerotolerant bacteria. The microbial communities are highly variable in particular individuals and compose of about 140 distinct families. The bronchial tree for instance contains a mean of 2000 bacterial genomes per cm2 surface. The harmful or potentially harmful bacteria are also detected routinely in respiratory specimens. The most significant are ''Moraxella catarrhalis'', ''Haemophilus inf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
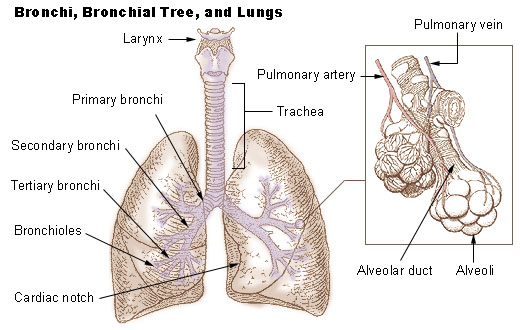
Lung
The lungs are the primary organs of the respiratory system in humans and most other animals, including some snails and a small number of fish. In mammals and most other vertebrates, two lungs are located near the backbone on either side of the heart. Their function in the respiratory system is to extract oxygen from the air and transfer it into the bloodstream, and to release carbon dioxide from the bloodstream into the atmosphere, in a process of gas exchange. Respiration is driven by different muscular systems in different species. Mammals, reptiles and birds use their different muscles to support and foster breathing. In earlier tetrapods, air was driven into the lungs by the pharyngeal muscles via buccal pumping, a mechanism still seen in amphibians. In humans, the main muscle of respiration that drives breathing is the diaphragm. The lungs also provide airflow that makes vocal sounds including human speech possible. Humans have two lungs, one on the left and on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Moraxella Catarrhalis
''Moraxella catarrhalis'' is a fastidious, nonmotile, Gram-negative, aerobic, oxidase-positive diplococcus that can cause infections of the respiratory system, middle ear, eye, central nervous system, and joints of humans. It causes the infection of the host cell by sticking to the host cell using trimeric autotransporter adhesins. Epidemiology ''Moraxella catarrhalis'' is a human pathogen with an affinity for the human upper respiratory tract. Other primates, such as macaques, might become infected by this bacterium. History ''Moraxella catarrhalis'' was previously placed in a separate genus named ''Branhamella''. The rationale for this was that other members of the genus ''Moraxella'' are rod-shaped and rarely caused infections in humans. However, results from DNA hybridization studies and 16S rRNA sequence comparisons were used to justify inclusion of the species ''M. catarrhalis'' in the genus ''Moraxella''. As a consequence, the name ''Moraxella catarrhalis'' is cur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microbe-associated Molecular Patterns
Pathogen-associated molecular patterns (PAMPs) are small molecular motifs conserved within a class of microbes. They are recognized by toll-like receptors (TLRs) and other pattern recognition receptors (PRRs) in both plants and animals. A vast array of different types of molecules can serve as PAMPs, including glycans and glycoconjugates. PAMPs activate innate immune responses, protecting the host from infection, by identifying some conserved nonself molecules. Bacterial lipopolysaccharides (LPSs), endotoxins found on the cell membranes of gram-negative bacteria, are considered to be the prototypical class of PAMPs. LPSs are specifically recognised by TLR4, a recognition receptor of the innate immune system. Other PAMPs include bacterial flagellin (recognized by TLR5), lipoteichoic acid from gram-positive bacteria (recognized by TLR2), peptidoglycan (recognized by TLR2), and nucleic acid variants normally associated with viruses, such as double-stranded RNA (dsRNA), recognized by T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) is a protein complex that controls transcription of DNA, cytokine production and cell survival. NF-κB is found in almost all animal cell types and is involved in cellular responses to stimuli such as stress, cytokines, free radicals, heavy metals, ultraviolet irradiation, oxidized LDL, and bacterial or viral antigens. NF-κB plays a key role in regulating the immune response to infection. Incorrect regulation of NF-κB has been linked to cancer, inflammatory and autoimmune diseases, septic shock, viral infection, and improper immune development. NF-κB has also been implicated in processes of synaptic plasticity and memory. Discovery NF-κB was discovered by Ranjan Sen in the lab of Nobel laureate David Baltimore via its interaction with an 11-base pair sequence in the immunoglobulin light-chain enhancer in B cells. Later work by Alexander Poltorak and Bruno Lemaitre in mice and ''Drosophila'' frui ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RIG-I-like Receptor
RIG-like receptors (retinoic acid-inducible gene-I-like receptors, RLRs) are a type of intracellular pattern recognition receptor involved in the recognition of viruses by the innate immune system. RIG-I (retinoic-acid inducible gene or DDX58) is the best characterized receptor within the RIG-I like receptor (RLR) family. Together with MDA5 (melanoma differentiation-associated 5) and LGP2 (laboratory of genetics and physiology 2), this family of cytoplasmic pattern recognition receptors (PRRs) are sentinels for intracellular viral RNA that is a product of viral infection. The RLR receptors provide frontline defence against viral infections in most tissues. RLR ligands The RIG-I receptor prefers to bind short (2000 bp), such as the replicative form of picornavirus RNA that is found in picornavirus-infected cells. LGP2 binds to blunt-ended double-stranded RNA of variable length, and also to RNA-bound MDA5 to regulate filament formation. The latter is linked to LGP2's recognition of p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NOD-like Receptor
The nucleotide-binding oligomerization domain-like receptors, or NOD-like receptors (NLRs) (also known as nucleotide-binding leucine-rich repeat receptors), are intracellular sensors of pathogen-associated molecular patterns (PAMPs) that enter the cell via phagocytosis or pores, and damage-associated molecular patterns (DAMPs) that are associated with cell stress. They are types of pattern recognition receptors (PRRs), and play key roles in the regulation of innate immune response. NLRs can cooperate with toll-like receptors (TLRs) and regulate inflammatory and apoptotic response. They are found in lymphocytes, macrophages, dendritic cells and also in non-immune cells, for example in epithelium. NLRs are highly conserved through evolution. Their homologs have been discovered in many different animal species (APAF1) and also in the plant kingdom ( disease-resistance R protein). Structure NLRs contain 3 domains – central NACHT (NOD or NBD – nucleotide-binding domain) domai ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Toll Like Receptors
Toll-like receptors (TLRs) are a class of proteins that play a key role in the innate immune system. They are single-pass membrane-spanning receptors usually expressed on sentinel cells such as macrophages and dendritic cells, that recognize structurally conserved molecules derived from microbes. Once these microbes have reached physical barriers such as the skin or intestinal tract mucosa, they are recognized by TLRs, which activate immune cell responses. The TLRs include TLR1, TLR2, TLR3, TLR4, TLR5, TLR6, TLR7, TLR8, TLR9, TLR10, TLR11, TLR12, and TLR13. Humans lack genes for TLR11, TLR12 and TLR13 and mice lack a functional gene for TLR10. TLR1, TLR2, TLR4, TLR5, TLR6, and TLR10 are located on the cell membrane, whereas TLR3, TLR7, TLR8, and TLR9 are located in intracellular vesicles (because they are sensors of nucleic acids). TLRs received their name from their similarity to the protein coded by the toll gene. Function The ability of the immune system to recognize mole ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Commensals Vs Pathogens Mechanism
Commensalism is a long-term biological interaction ( symbiosis) in which members of one species gain benefits while those of the other species neither benefit nor are harmed. This is in contrast with mutualism, in which both organisms benefit from each other; amensalism, where one is harmed while the other is unaffected; parasitism, where one is harmed and the other benefits, and parasitoidism, which is similar to parasitism but the parasitoid has a free-living state and instead of just harming its host, it eventually ends up killing it. The commensal (the species that benefits from the association) may obtain nutrients, shelter, support, or locomotion from the host species, which is substantially unaffected. The commensal relation is often between a larger host and a smaller commensal; the host organism is unmodified, whereas the commensal species may show great structural adaptation consistent with its habits, as in the remoras that ride attached to sharks and other fishes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aspergillus
'''' () is a genus consisting of several hundred mold species found in various climates worldwide. ''Aspergillus'' was first catalogued in 1729 by the Italian priest and biologist Pier Antonio Micheli. Viewing the fungi under a microscope, Micheli was reminded of the shape of an ''aspergillum'' (holy water sprinkler), from Latin ''spargere'' (to sprinkle), and named the genus accordingly. Aspergillum is an asexual spore-forming structure common to all ''Aspergillus'' species; around one-third of species are also known to have a sexual stage. While some species of ''Aspergillus'' are known to cause fungal infections, others are of commercial importance. Taxonomy Species ''Aspergillus'' consists of 837 species of fungi. Growth and distribution ''Aspergillus'' is defined as a group of conidial fungi—that is, fungi in an asexual state. Some of them, however, are known to have a teleomorph (sexual state) in the Ascomycota. With DNA evidence, all members of the genus '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Saccharomyces
''Saccharomyces'' is a genus of fungi that includes many species of yeasts. ''Saccharomyces'' is from Greek σάκχαρον (sugar) and μύκης (fungus) and means ''sugar fungus''. Many members of this genus are considered very important in food production. It is known as the brewer's yeast or baker's yeast. They are unicellular and saprotrophic fungi. One example is ''Saccharomyces cerevisiae'', which is used in making bread, wine, and beer, and for human and animal health. Other members of this genus include the wild yeast ''Saccharomyces paradoxus'' that is the closest relative to ''S. cerevisiae'', ''Saccharomyces bayanus'', used in making wine, and ''Saccharomyces cerevisiae'' var. ''boulardii'', used in medicine. Morphology Colonies of ''Saccharomyces'' grow rapidly and mature in three days. They are flat, smooth, moist, glistening or dull, and cream in color. The inability to use nitrate and ability to ferment various carbohydrates are typical characteristics of ''Sacc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Malassezia
''Malassezia'' (formerly known as ''Pityrosporum'') is a genus of fungi. It is the sole genus in family Malasseziaceae, which is the only family in order Malasseziales, itself the single member of class Malasseziomycetes. ''Malassezia'' species are naturally found on the skin surfaces of many animals, including humans. In occasional opportunistic infections, some species can cause hypopigmentation or hyperpigmentation on the trunk and other locations in humans. Allergy tests for these fungi are available. Systematics Due to progressive changes in their nomenclature, some confusion exists about the naming and classification of ''Malassezia'' yeast species. Work on these yeasts has been complicated because they require specific growth media and grow very slowly in laboratory culture. ''Malassezia'' were originally identified by the French scientist Louis-Charles Malassez in the late nineteenth century. Raymond Sabouraud identified a dandruff-causing organism in 1904 and called i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |