|
List Of Gene Prediction Software
This is a list of software tools and web portals used for gene prediction. See also * Gene prediction * List of RNA structure prediction software * Comparison of software for molecular mechanics modeling References {{Reflist, 2 Prediction A prediction (Latin ''præ-'', "before," and ''dicere'', "to say"), or forecast, is a statement about a future event or data. They are often, but not always, based upon experience or knowledge. There is no universal agreement about the exact ... gene prediction software ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Software
Software is a set of computer programs and associated software documentation, documentation and data (computing), data. This is in contrast to Computer hardware, hardware, from which the system is built and which actually performs the work. At the low level language, lowest programming level, executable code consists of Machine code, machine language instructions supported by an individual Microprocessor, processor—typically a central processing unit (CPU) or a graphics processing unit (GPU). Machine language consists of groups of Binary number, binary values signifying Instruction set architecture, processor instructions that change the state of the computer from its preceding state. For example, an instruction may change the value stored in a particular storage location in the computer—an effect that is not directly observable to the user. An instruction System call, may also invoke one of many Input/output, input or output operations, for example displaying some text on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GeneMark
GeneMark is a generic name for a family of ab initio gene prediction programs developed at the Georgia Institute of Technology in Atlanta. Developed in 1993, original GeneMark was used in 1995 as a primary gene prediction tool for annotation of the first completely sequenced bacterial genome of '' Haemophilus influenzae'', and in 1996 for the first archaeal genome of ''Methanococcus jannaschii''. The algorithm introduced inhomogeneous three-periodic Markov chain models of protein-coding DNA sequence that became standard in gene prediction as well as Bayesian approach to gene prediction in two DNA strands simultaneously. Species specific parameters of the models were estimated from training sets of sequences of known type (protein-coding and non-coding). The major step of the algorithm computes for a given DNA fragment posterior probabilities of either being "protein-coding" (carrying genetic code) in each of six possible reading frames (including three frames in complementary ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparison Of Software For Molecular Mechanics Modeling
This is a list of computer programs that are predominantly used for molecular mechanics calculations. See also * Car–Parrinello molecular dynamics *Comparison of force-field implementations * Comparison of nucleic acid simulation software *List of molecular graphics systems * List of protein structure prediction software *List of quantum chemistry and solid-state physics software *List of software for Monte Carlo molecular modeling *List of software for nanostructures modeling This is a list of computer programs that are used to model nanostructures at the levels of classical mechanics and quantum mechanics. Furiousatoms- a powerful software for molecular modelling and visualization * Aionics.io - a powerful platform ... * Molecular design software * Molecular dynamics * Molecular modeling on GPUs * Molecule editor Notes and references External linksSINCRIS [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of RNA Structure Prediction Software
This list of RNA structure prediction software is a compilation of software tools and web portals used for RNA structure prediction. Single sequence secondary structure prediction. Single sequence tertiary structure prediction Comparative methods The single sequence methods mentioned above have a difficult job detecting a small sample of reasonable secondary structures from a large space of possible structures. A good way to reduce the size of the space is to use evolutionary approaches. Structures that have been conserved by evolution are far more likely to be the functional form. The methods below use this approach. RNA solvent accessibility prediction Intermolecular interactions: RNA-RNA Many ncRNAs function by binding to other RNAs. For example, miRNAs regulate protein coding gene expression by binding to 3' UTRs, small nucleolar RNAs guide post-transcriptional modifications by binding to rRNA, U4 spliceosomal RNA and U6 spliceosomal RNA bind to each other forming pa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Prediction
In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. This includes protein-coding genes as well as RNA genes, but may also include prediction of other functional elements such as regulatory regions. Gene finding is one of the first and most important steps in understanding the genome of a species once it has been sequenced. In its earliest days, "gene finding" was based on painstaking experimentation on living cells and organisms. Statistical analysis of the rates of homologous recombination of several different genes could determine their order on a certain chromosome, and information from many such experiments could be combined to create a genetic map specifying the rough location of known genes relative to each other. Today, with comprehensive genome sequence and powerful computational resources at the disposal of the research community, gene finding has been redefined as a largely compu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Primary Transcript
A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs are modified in preparation for translation. For example, a precursor mRNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after processing. Pre-mRNA is synthesized from a DNA template in the cell nucleus by transcription. Pre-mRNA comprises the bulk of heterogeneous nuclear RNA (hnRNA). Once pre-mRNA has been completely processed, it is termed "mature messenger RNA", or simply " messenger RNA". The term hnRNA is often used as a synonym for pre-mRNA, although, in the strict sense, hnRNA may include nuclear RNA transcripts that do not end up as cytoplasmic mRNA. There are several steps contributing to the production of primary transcripts. All these steps involve a series of interactions to initiate and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes (e.g. MS2) and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm. Bacteriophages are among the most common and diverse entities in the biosphere. Bacteriophages are ubiquitous viruses, found wherever bacteria exist. It is estimated there are more than 1031 bacteriophages on the planet, more than every other organism on Earth, including bacteria, combined. Viruses are the most abundant biological entity in the water column of the world's oceans, and the second largest component of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
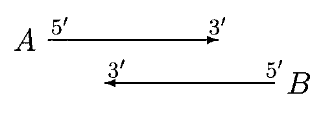
Open Reading Frame
In molecular biology, open reading frames (ORFs) are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a Prokaryote, prokaryotic DNA sequence, where only one of the #Six-frame translation, six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by Transcription (biology), transcription of the DNA and its subsequent interaction with the ribosome in Translation (biology), translation). Such an ORF may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription terminator, transcription termination site is located after the ORF, beyond the Translation (biology), translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neural Network
A neural network is a network or neural circuit, circuit of biological neurons, or, in a modern sense, an artificial neural network, composed of artificial neurons or nodes. Thus, a neural network is either a biological neural network, made up of biological neurons, or an artificial neural network, used for solving artificial intelligence (AI) problems. The connections of the biological neuron are modeled in artificial neural networks as weights between nodes. A positive weight reflects an excitatory connection, while negative values mean inhibitory connections. All inputs are modified by a weight and summed. This activity is referred to as a linear combination. Finally, an activation function controls the amplitude of the output. For example, an acceptable range of output is usually between 0 and 1, or it could be −1 and 1. These artificial networks may be used for predictive modeling, adaptive control and applications where they can be trained via a dataset. Self-learning re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Support-vector Machine
In machine learning, support vector machines (SVMs, also support vector networks) are supervised learning models with associated learning algorithms that analyze data for classification and regression analysis. Developed at AT&T Bell Laboratories by Vladimir Vapnik with colleagues (Boser et al., 1992, Guyon et al., 1993, Cortes and Vapnik, 1995, Vapnik et al., 1997) SVMs are one of the most robust prediction methods, being based on statistical learning frameworks or VC theory proposed by Vapnik (1982, 1995) and Chervonenkis (1974). Given a set of training examples, each marked as belonging to one of two categories, an SVM training algorithm builds a model that assigns new examples to one category or the other, making it a non-probabilistic binary linear classifier (although methods such as Platt scaling exist to use SVM in a probabilistic classification setting). SVM maps training examples to points in space so as to maximise the width of the gap between the two categorie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GLIMMER
In bioinformatics, GLIMMER (Gene Locator and Interpolated Markov ModelER) is used to find genes in prokaryotic DNA. "It is effective at finding genes in bacteria, archea, viruses, typically finding 98-99% of all relatively long protein coding genes". GLIMMER was the first system that used the interpolated Markov model to identify coding regions. The GLIMMER software is open source and is maintained by Steven Salzberg, Art Delcher, and their colleagues at the ''Center for Computational Biology'' at Johns Hopkins University. The original GLIMMER algorithms and software were designed by Art Delcher, Simon Kasif and Steven Salzberg and applied to bacterial genome annotation in collaboration with Owen White. Versions GLIMMER 1.0 First Version of GLIMMER "i.e., GLIMMER 1.0" was released in 1998 and it was published in the paper ''Microbial gene identification using interpolated Markov model''. Markov models were used to identify microbial genes in GLIMMER 1.0. GLIMMER considers the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GENSCAN
In bioinformatics, GENSCAN is a program to identify complete gene structures in genomic DNA. It is a G HMM-based program that can be used to predict the location of genes and their exon-intron boundaries in genomic sequences from a variety of organisms. The GENSCAN Web server can be found at MIT. GENSCAN was developed by Christopher Burge in the research group of Samuel Karlin at Stanford University. History In 2001, the world of human gene prediction entered into Comparative genomics. This resulted in the development of a program called TWINSCAN as an adaptation of GENSCAN with higher accuracy. Other programs like N-SCAN were later developed by further adapting the GHMM model. As of 2002, GENSCAN remained a popular tool in bioinformatics, becoming a standard feature for genomes released on University of California Santa Cruz and Ensembl Genome browser. Implementation Genomic Model The primary goal when developing a genomic sequence model for GENSCAN was to identify both ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |