|
List Of EC Numbers (EC 5)
This list contains a list of EC numbers for the fifth group, EC 5, isomerases, placed in numerical order as determined by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. All official information is tabulated at the website of the committee. The database is developed and maintained by Andrew McDonald. EC 5.1: Epimerases and racemases EC 5.1.1: Acting on Amino acids and Derivatives * : alanine racemase * : methionine racemase * : glutamate racemase * : proline racemase * : lysine racemase * : threonine racemase * : diaminopimelate epimerase * : 4-hydroxyproline epimerase * : arginine racemase * : amino-acid racemase * : phenylalanine racemase (ATP-hydrolysing) * : ornithine racemase * : aspartate racemase * : nocardicin-A epimerase * : 2-aminohexano-6-lactam racemase * : protein-serine epimerase * : isopenicillin-N epimerase * : serine racemase * : L-Ala-D/L-Glu epimerase * * : isoleucine 2-epimerase * * : 4-hydroxypr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme Commission Number
The Enzyme Commission number (EC number) is a numerical classification scheme for enzymes, based on the chemical reactions they catalyze. As a system of enzyme nomenclature, every EC number is associated with a recommended name for the corresponding enzyme-catalyzed reaction. EC numbers do not specify enzymes but enzyme-catalyzed reactions. If different enzymes (for instance from different organisms) catalyze the same reaction, then they receive the same EC number. Furthermore, through convergent evolution, completely different protein folds can catalyze an identical reaction (these are sometimes called non-homologous isofunctional enzymes) and therefore would be assigned the same EC number. By contrast, UniProt identifiers uniquely specify a protein by its amino acid sequence. Format of number Every enzyme code consists of the letters "EC" followed by four numbers separated by periods. Those numbers represent a progressively finer classification of the enzyme. Preliminary EC ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
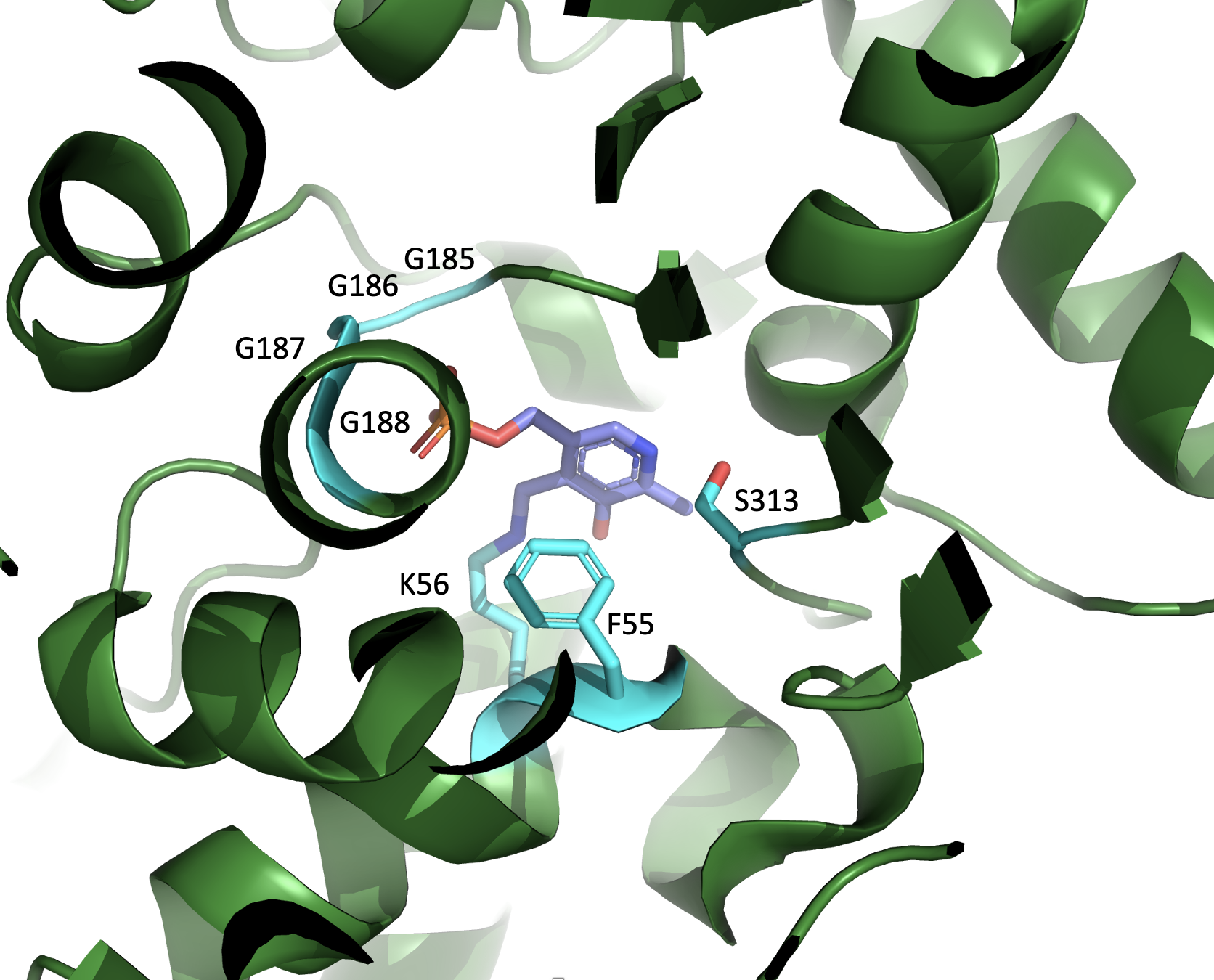
Aspartate Racemase
In enzymology, an aspartate racemase () is an enzyme that catalyzes the following chemical reaction: :L-aspartate \rightleftharpoons D-aspartate This enzyme belongs to the family of isomerases, specifically those racemases and epimerases acting on amino acids and amino acid derivatives, including glutamate racemase, proline racemase, and diaminopimelate epimerase. The systematic name of this enzyme class is aspartate racemase. Other names in common use include D-aspartate racemase, and McyF. Discovery Aspartate racemase was first discovered in the gram-positive bacteria ''Streptococcus faecalis'' by Lamont ''et al''. in 1972. It was then determined that aspartate racemase also racemizes L-alanine around half as quickly as it does L-aspartate, but does not show racemase activity in the presence of L-glutamate. Structure The crystallographic structure of bacterial aspartate racemase has been solved in ''Pyrococcus horikoshii OT3'', ''Escherichia coli'', ''Microcystis aeru ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carbohydrates
In organic chemistry, a carbohydrate () is a biomolecule consisting of carbon (C), hydrogen (H) and oxygen (O) atoms, usually with a hydrogen–oxygen atom ratio of 2:1 (as in water) and thus with the empirical formula (where ''m'' may or may not be different from ''n''), which does not mean the H has covalent bonds with O (for example with , H has a covalent bond with C but not with O). However, not all carbohydrates conform to this precise stoichiometric definition (e.g., uronic acids, deoxy-sugars such as fucose), nor are all chemicals that do conform to this definition automatically classified as carbohydrates (e.g. formaldehyde and acetic acid). The term is most common in biochemistry, where it is a synonym of saccharide (), a group that includes sugars, starch, and cellulose. The saccharides are divided into four chemical groups: monosaccharides, disaccharides, oligosaccharides, and polysaccharides. Monosaccharides and disaccharides, the smallest (lower molecular weight) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isocitrate Epimerase
In enzymology, an isocitrate epimerase is classified as follows: . This number indicates that it is an isomerase, specifically a racemase or epimerase that acts on hydroxy acids and their derivatives, namely isocitrate. Isocitrate epimerase specifically catalyzes the reversible reaction: :(1''R'',2''S'')-1-hydroxypropane-1,2,3-tricarboxylate ↔ (1''S'',2''S'')-1-hydroxypropane-1,2,3-tricarboxylate which can also be described as :D-threo-isocitrate ↔ D-erythro-isocitrate History Isocitrate epimerase was originally isolated from the fungal cell-free extract of ''Penicillium purpurogenum'' '','' where it was discovered due to the excess accumulation of L-alloisocitric acid (D-erythro-isocitrate)—a diastereomer of isocitrate previously not seen in nature. In order to accumulate L-alloisocitric acid as a fermentation product, ''P. purpurogenum'' needed to be grown on citrate supplemented nutrient agar. During this fermentation it was found that the fermentation yield of L-allois ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tartrate Epimerase
In enzymology, a tartrate epimerase () is an enzyme that catalyzes the chemical reaction :(R,R)-tartrate \rightleftharpoons meso-tartrate Hence, this enzyme has one substrate, (R,R)-tartrate, and one product, meso-tartrate. This enzyme belongs to the family of isomerases, specifically those racemases and epimerases acting on hydroxy acids and derivatives. The systematic name of this enzyme class is tartrate epimerase. This enzyme is also called tartaric racemase. This enzyme participates in glyoxylate and dicarboxylate In organic chemistry, a carboxylate is the conjugate base of a carboxylic acid, (or ). It is an ion with negative charge. Carboxylate salts are salts that have the general formula , where M is a metal and ''n'' is 1, 2,...; ''carboxylat ... metabolism. References * EC 5.1.2 Enzymes of unknown structure {{isomerase-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Acetoin Racemase
In enzymology, an acetoin racemase () is an enzyme that catalyzes the chemical reaction :(S)-acetoin \rightleftharpoons (R)-acetoin This enzyme belongs to the family of isomerases, specifically those racemases and epimerases acting on hydroxy acids and derivatives. The systematic name of this enzyme class is acetoin racemase. This enzyme is also called acetylmethylcarbinol racemase. This enzyme participates in butanoate metabolism Butyric acid (; from grc, βούτῡρον, meaning "butter"), also known under the systematic name butanoic acid, is a straight-chain alkyl carboxylic acid with the chemical formula CH3CH2CH2CO2H. It is an oily, colorless liquid with an unple .... References * EC 5.1.2 Enzymes of unknown structure {{isomerase-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
3-hydroxybutyryl-CoA Epimerase
In enzymology, a 3-hydroxybutyryl-CoA epimerase () is an enzyme that catalyzes the chemical reaction :(S)-3-hydroxybutanoyl-CoA \rightleftharpoons (R)-3-hydroxybutanoyl-CoA Hence, this enzyme has one substrate, (S)-3-hydroxybutanoyl-CoA, and one product, (R)-3-hydroxybutanoyl-CoA. This enzyme belongs to the family of isomerases, specifically those racemases and epimerases acting on hydroxy acids and derivatives. The systematic name of this enzyme class is 3-hydroxybutanoyl-CoA 3-epimerase. Other names in common use include 3-hydroxybutyryl coenzyme A epimerase, and 3-hydroxyacyl-CoA epimerase. This enzyme participates in fatty acid metabolism and butanoate metabolism. Structural studies As of late 2007, four structures A structure is an arrangement and organization of interrelated elements in a material object or system, or the object or system so organized. Material structures include man-made objects such as buildings and machines and natural objects such as ... have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mandelate Racemase
Mandelate racemase () is a bacterial enzyme which catalyzes the interconversion of the enantiomers of mandelate via an enol intermediate. This enzyme catalyses the following chemical reaction : (S)-mandelate \rightleftharpoons (R)-mandelate It is a member of the enolase superfamily of enzymes, along with muconate lactonizing enzyme and enolase Phosphopyruvate hydratase, usually known as enolase, is a metalloenzyme () that catalyses the conversion of 2-phosphoglycerate (2-PG) to phosphoenolpyruvate (PEP), the ninth and penultimate step of glycolysis. The chemical reaction is: :2-p .... References External links * EC 5.1.2 {{Enzyme-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lactate Racemase
The lactate racemase enzyme (Lar) () interconverts the D- and L-enantiomers of lactic acid. It is classified under the isomerase, racemase, epimerase, and enzyme acting on hydroxyl acids and derivatives classes of enzymes. It is found in certain halophilic archaea, such as ''Haloarcula marismortui'', and in a few species of bacteria, such as several ''Lactobacillus'' species (which produce D- and L- lactate) including ''Lactobacillus sakei'', ''Lactobacillus curvatus'', and ''Lactobacillus plantarum'', as well as in non-lactic acid bacteria such as ''Clostridium beijerinckii''. The gene encoding lactate racemase in ''L. plantarum'' was identified as ''larA'' and shown to be associated with a widespread maturation system involving ''larB'', ''larC1'', ''larC2'', and ''larE''. The optimal pH for its activity is 5.8-6.2 in ''L. sakei''. Structure and properties The molecular weight of lactate racemase differs in the various organisms in which it has been found, ranging from 25,000 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydroxy Acids
α-Hydroxy acids, or alpha hydroxy acids (AHAs), are a class of chemical compounds that consist of a carboxylic acid with a hydroxyl group substituent on the adjacent (alpha) carbon. Prominent examples are glycolic acid, lactic acid, mandelic acid and citric acid. Although these compounds are related to the ordinary carboxylic acids and are therefore weak acids, their chemical structure allows for the formation of an internal hydrogen bond between the hydrogen at the hydroxyl group and one of the oxygen atoms of the carboxylic group. The net effect is an increase in acidity. For example, the pKa of lactic acid is 3.86, while that of the unsubstituted propionic acid is 4.87; a full pKa unit difference means that lactic acid is ten times stronger than propionic acid. Industrial applications Feed additives 2-Hydroxy-4-(methylthio)butyric acid is produced commercially as a racemic mixture to substitute for methionine in animal feed. In nature, the same compound is an interm ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine Racemase
Serine racemase (SR, ) is the first racemase enzyme in human biology to be identified. This enzyme converts L- serine to its enantiomer form, D-serine. D-serine acts as a neuronal signaling molecule by activating NMDA receptors in the brain. Since NMDA receptors Dysfunction has been suggested as one of the promising hypotheses for the pathophysiology of schizophrenia, it has been shown that underexpression of this enzyme is an indicator, especially for the paranoid subtype. Treatment of schizophrenia with D-serine has been shown to play some role in ameliorating some symptoms. In humans, the serine racemase protein is encoded by the ''SRR'' gene. Serine racemase may have evolved from L-thre-hydroxyaspartate (L-THA) eliminase and served as the precursor to aspartate racemase. Mammalian serine racemase is a pyridoxal 5'-phosphate dependent enzyme that catalyzes both the racemization of L-serine to D-serine and also the elimination of water from L-serine, generating pyruvate and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isopenicillin-N Epimerase
In enzymology, an isopenicillin N epimerase () is an enzyme that catalyzes the chemical reaction :isopenicillin N \rightleftharpoons penicillin N Hence, this enzyme has one substrate, isopenicillin N, and one product, penicillin N. This enzyme belongs to the family of isomerases, specifically those racemases and epimerases acting on amino acids and derivatives. The systematic name A systematic name is a name given in a systematic way to one unique group, organism, object or chemical substance, out of a specific population or collection. Systematic names are usually part of a nomenclature. A semisystematic name or semitrivial ... of this enzyme class is penicillin N 5-amino-5-carboxypentanoyl-epimerase. This enzyme participates in penicillin and cephalosporin biosynthesis. References * * * * EC 5.1.1 Enzymes of unknown structure {{isomerase-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |