|
LiMETER
LiMETER stands for light-inducible membrane-tethered peripheral endoplasmic reticulum (ER). LiMETER is an optogenetics tool designed to reversibly label cortical ER or the apposition between plasma membrane (PM) and endoplasmic reticulum (ER) membranes (termed as ER-PM junctions). Design The ER luminal domain of LiMETER contains a signal peptide and the transmembrane domain derived from STIM1, with GFP placed in between as a reporter. STIM1 is an ER-resident calcium sensor protein responsible for sensing calcium changes in internal calcium stores and communicate with ORAI calcium channels in the plasma membrane. The cytoplasmic region of LiMETER contains a flexible linker and a genetically encoded lightswitch LOV2 domain (light oxygen voltage-sensing domain, residues 404–546) derived from ''Avena sativa'' phototropin 1, followed by a C-terminal PM-targeting polybasic tail that associates with negative charged phosphoinositides Phosphatidylinositol (or Inositol Phospholip ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Membrane Contact Site
Membrane contact sites (MCS) are close appositions between two organelles. Ultrastructural studies typically reveal an intermembrane distance in the order of the size of a single protein, as small as 10 nm or wider, with no clear upper limit. These zones of apposition are highly conserved in evolution. These sites are thought to be important to facilitate signalling, and they promote the passage of small molecules, including ions, lipids and (discovered later) reactive oxygen species. MCS are important in the function of the endoplasmic reticulum (ER), since this is the major site of lipid synthesis within cells. The ER makes close contact with many organelles, including mitochondria, Golgi, endosomes, lysosomes, peroxisomes, chloroplasts and the plasma membrane. Both mitochondria and sorting endosomes undergo major rearrangements leading to fission where they contact the ER. Sites of close apposition can also form between most of these organelles most pairwise combinations. Fi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Optogenetics
Optogenetics is a biological technique to control the activity of neurons or other cell types with light. This is achieved by expression of light-sensitive ion channels, pumps or enzymes specifically in the target cells. On the level of individual cells, light-activated enzymes and transcription factors allow precise control of biochemical signaling pathways. In systems neuroscience, the ability to control the activity of a genetically defined set of neurons has been used to understand their contribution to decision making, learning, fear memory, mating, addiction, feeding, and locomotion. In a first medical application of optogenetic technology, vision was partially restored in a blind patient. Optogenetic techniques have also been introduced to map the functional connectivity of the brain''.'' By altering the activity of genetically labelled neurons with light and using imaging and electrophysiology techniques to record the activity of other cells, researchers can identify ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
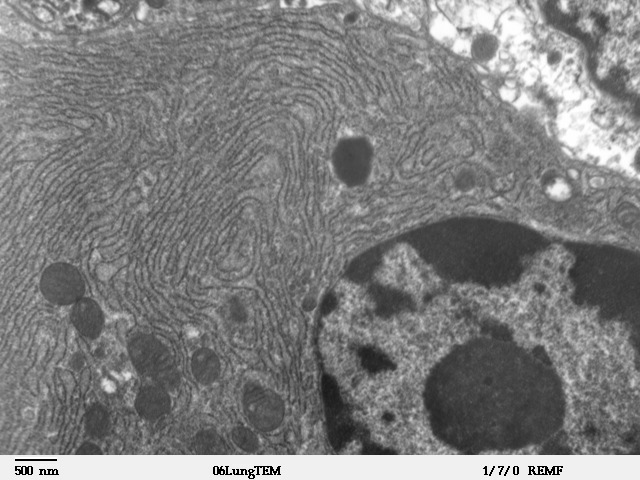
Endoplasmic Reticulum
The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum (RER), and smooth endoplasmic reticulum (SER). The endoplasmic reticulum is found in most eukaryotic cells and forms an interconnected network of flattened, membrane-enclosed sacs known as cisternae (in the RER), and tubular structures in the SER. The membranes of the ER are continuous with the outer nuclear membrane. The endoplasmic reticulum is not found in red blood cells, or spermatozoa. The two types of ER share many of the same proteins and engage in certain common activities such as the synthesis of certain lipids and cholesterol. Different types of cells contain different ratios of the two types of ER depending on the activities of the cell. RER is found mainly toward the nucleus of cell and SER towards the cell membrane or plasma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signal Peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16-30 amino acids long) present at the N-terminus (or occasionally nonclassically at the C-terminus or internally) of most newly synthesized proteins that are destined toward the secretory pathway. These proteins include those that reside either inside certain organelles (the endoplasmic reticulum, Golgi or endosomes), secreted from the cell, or inserted into most cellular membranes. Although most type I membrane-bound proteins have signal peptides, the majority of type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a signal sequence except that it is not cleaved. They are a kind of target peptide. Function (translocation) Signal peptides function to prompt a cell to translo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmembrane Domain
A transmembrane domain (TMD) is a membrane-spanning protein domain. TMDs generally adopt an alpha helix topological conformation, although some TMDs such as those in porins can adopt a different conformation. Because the interior of the lipid bilayer is hydrophobic, the amino acid residues in TMDs are often hydrophobic, although proteins such as membrane pumps and ion channels can contain polar residues. TMDs vary greatly in length, sequence, and hydrophobicity, adopting organelle-specific properties. Functions of transmembrane domains Transmembrane domains are known to perform a variety of functions. These include: * Anchoring transmembrane proteins to the membrane. *Facilitating molecular transport of molecules such as ions and proteins across biological membranes; usually hydrophilic residues and binding sites in the TMDs help in this process. *Signal transduction across the membrane; many transmembrane proteins, such as G protein-coupled receptors, receive extracellular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
STIM1
Stromal interaction molecule 1 is a protein that in humans is encoded by the ''STIM1'' gene. STIM1 has a single transmembrane domain, and is localized to the endoplasmic reticulum, and to a lesser extent to the plasma membrane. Even though the protein has been identified earlier, its function was unknown until recently. In 2005, it was discovered that STIM1 functions as a calcium sensor in the endoplasmic reticulum. Upon activation of the IP3 receptor, the calcium concentration in the endoplasmic reticulum decreases, which is sensed by STIM1, via its EF hand domain. STIM1 activates the "store-operated" ORAI1 calcium ion channels in the plasma membrane, via intracellular STIM1 movement, clustering under plasma membrane and protein interaction with ORAI isoforms. STIM1-mediated calcium entry is required for thrombin-induced disassembly of VE-cadherin adherens junctions. 2-Aminoethoxydiphenyl borate (2-APB) and 4-chloro-3-ethylphenol (4-CEP) cause STIM1 clustering in a cell and preven ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Green Fluorescent Protein
The green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range. The label ''GFP'' traditionally refers to the protein first isolated from the jellyfish ''Aequorea victoria'' and is sometimes called ''avGFP''. However, GFPs have been found in other organisms including corals, sea anemones, zoanithids, copepods and lancelets. The GFP from ''A. victoria'' has a major excitation peak at a wavelength of 395 nm and a minor one at 475 nm. Its emission peak is at 509 nm, which is in the lower green portion of the visible spectrum. The fluorescence quantum yield (QY) of GFP is 0.79. The GFP from the sea pansy (''Renilla reniformis'') has a single major excitation peak at 498 nm. GFP makes for an excellent tool in many forms of biology due to its ability to form an internal chromophore without requiring any accessory cofactors, gene products, or enzymes / substrates other than mo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ORAI1
Calcium release-activated calcium channel protein 1 is a calcium selective ion channel that in humans is encoded by the ''ORAI1'' gene. Orai channels play an important role in the activation of T-lymphocytes. The loss of function mutation of Orai1 causes severe combined immunodeficiency (SCID) in humans The mammalian orai family has two additional homologs, Orai2 and Orai3. Orai proteins share no homology with any other ion channel family of any other known proteins. They have 4 transmembrane domains and form hexamers. Structure and function Orai channels are activated upon the depletion of internal calcium stores, which is called the "store-operated" or the "capacitative" mechanism. They are molecular constituents of the "calcium release activated calcium currents" ( ICRAC). Upon activation of phospholipase C by various cell surface receptors, inositol trisphosphate is formed that releases calcium from the endoplasmic reticulum. The decreased calcium concentration in the endopl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Light-Oxygen-Voltage-sensing Domain
A Light-oxygen-voltage-sensing domain (LOV domain) is a protein sensor used by a large variety of higher plants, microalgae, fungi and bacteria to sense environmental conditions. In higher plants, they are used to control phototropism, chloroplast relocation, and stomatal opening, whereas in fungal organisms, they are used for adjusting the circadian temporal organization of the cells to the daily and seasonal periods. They are a subset of PAS domains. Chromophore Common to all LOV proteins is the blue-light sensitive flavin chromophore, which in the signaling state is covalently linked to the protein core via an adjacent cysteine residue. LOV domains are e.g. encountered in phototropins, which are blue-light-sensitive protein complexes regulating a great diversity of biological processes in higher plants as well as in micro-algae. Phototropins are composed of two LOV domains, each containing a non-covalently bound flavin mononucleotide (FMN) chromophore in its dark-state for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phototropin
Phototropins are photoreceptor proteins (more specifically, flavoproteins) that mediate phototropism responses in higher plants. Phototropins can be found throughout the leaves of a plant. Along with cryptochromes and phytochromes they allow plants to respond and alter their growth in response to the light environment. Phototropins may also be important for the opening of stomata and the movement of chloroplasts. These blue light receptors are seen across the entire green plant lineage. When Phototropins are hit with blue light, they induce a signal transduction pathway that alters the plant cells' functions in different ways. Phototropins are part of the phototropic sensory system in plants that causes various environmental responses in plants. Phototropins specifically will cause stems to bend towards light and stomata to open. Phototropins have been shown to impact the movement of chloroplast inside the cell. In addition phototropins mediate the first changes in stem elongation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphoinositides
Phosphatidylinositol (or Inositol Phospholipid) consists of a family of lipids as illustrated on the right, where red is x, blue is y, and black is z, in the context of independent variation, a class of the phosphatidylglycerides. In such molecules the isomer of the inositol group is assumed to be the myo- conformer unless otherwise stated. Typically phosphatidylinositols form a minor component on the cytosolic side of eukaryotic cell membranes. The phosphate group gives the molecules a negative charge at physiological pH. The form of phosphatidylinositol comprising the isomer ''muco''-inositol acts as a sensory receptor in the taste function of the sensory system. In this context it is often referred to as PtdIns, but that does not imply any molecular difference from phosphatidylinositols comprising the myo- conformers of inositol. The phosphatidylinositol can be phosphorylated to form phosphatidylinositol phosphate (PI-4-P, referred to as PIP in close context or inform ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cysteine
Cysteine (symbol Cys or C; ) is a semiessential proteinogenic amino acid with the formula . The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile. When present as a deprotonated catalytic residue, sometimes the symbol Cyz is used. The deprotonated form can generally be described by the symbol Cym as well. The thiol is susceptible to oxidation to give the disulfide derivative cystine, which serves an important structural role in many proteins. In this case, the symbol Cyx is sometimes used. When used as a food additive, it has the E number E920. Cysteine is encoded by the codons UGU and UGC. The sulfur-containing amino acids cysteine and methionine are more easily oxidized than the other amino acids. Structure Like other amino acids (not as a residue of a protein), cysteine exists as a zwitterion. Cysteine has chirality in the older / notation based on homology to - and -glyceraldehyde. In the newer ''R''/''S'' system of designating chi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |

.png)

