|
Lac Repressor
The ''lac'' repressor (LacI) is a DNA-binding protein that inhibits the expression of genes coding for proteins involved in the metabolism of lactose in bacteria. These genes are repressed when lactose is not available to the cell, ensuring that the bacterium only invests energy in the production of machinery necessary for uptake and utilization of lactose when lactose is present. When lactose becomes available, it is firstly converted into allolactose by β-Galactosidase (lacZ) in bacteria. The DNA binding ability of lac repressor bound with allolactose is inhibited due to allosteric regulation, thereby genes coding for proteins involved in lactose uptake and utilization can be expressed. Function The ''lac'' repressor (LacI) operates by a helix-turn-helix motif in its DNA-binding domain, binding base-specifically to the major groove of the operator region of the ''lac'' operon, with base contacts also made by residues of symmetry-related alpha helices, the "hinge" heli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LacI Dimer Structure Annotated
The ''lac'' repressor (LacI) is a DNA-binding protein that inhibits the Gene expression, expression of genes coding for proteins involved in the metabolism of lactose in bacteria. These genes are repressed when lactose is not available to the cell, ensuring that the bacterium only invests energy in the production of machinery necessary for uptake and utilization of lactose when lactose is present. When lactose becomes available, it is firstly converted into allolactose by Beta-galactosidase, β-Galactosidase (Lac operon, lacZ) in bacteria. The DNA binding ability of lac repressor bound with allolactose is inhibited due to allosteric regulation, thereby genes coding for proteins involved in lactose uptake and utilization can be expressed. Function The ''lac'' repressor (LacI) operates by a helix-turn-helix Structural motif, motif in its DNA-binding domain, binding base-specifically to the major groove of the Operon, operator region of the lac operon, ''lac'' operon, with base co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides. RNAP produces RNA that, functionally, is either for protein coding, i.e. messenger RNA (mRNA); or n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Dynamics
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics. Because molecular systems typically consist of a vast number of particles, it is impossible to determine the properties of such complex systems analytically; MD simulation circumvents this problem by using numerical methods. However, long MD simulations are mathematically ill-conditioned, generating cumulative errors in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Escherichia Coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escherichia'' that is commonly found in the lower intestine of warm-blooded organisms. Most ''E. coli'' strains are harmless, but some serotypes ( EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains do not cause disease in humans and are part of the normal microbiota of the gut; such strains are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of ''E. coli'' benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between ''E. col ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
In Vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, and plants, as opposed to a tissue extract or dead organism. This is not to be confused with experiments done ''in vitro'' ("within the glass"), i.e., in a laboratory environment using test tubes, Petri dishes, etc. Examples of investigations ''in vivo'' include: the pathogenesis of disease by comparing the effects of bacterial infection with the effects of purified bacterial toxins; the development of non-antibiotics, antiviral drugs, and new drugs generally; and new surgical procedures. Consequently, animal testing and clinical trials are major elements of ''in vivo'' research. ''In vivo'' testing is often employed over ''in vitro'' because it is better suited for observing the overall effects of an experiment on a living subject. In dr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Diffusion
Diffusion is the net movement of anything (for example, atoms, ions, molecules, energy) generally from a region of higher concentration to a region of lower concentration. Diffusion is driven by a gradient in Gibbs free energy or chemical potential. It is possible to diffuse "uphill" from a region of lower concentration to a region of higher concentration, like in spinodal decomposition. The concept of diffusion is widely used in many fields, including physics (particle diffusion), chemistry, biology, sociology, economics, and finance (diffusion of people, ideas, and price values). The central idea of diffusion, however, is common to all of these: a substance or collection undergoing diffusion spreads out from a point or location at which there is a higher concentration of that substance or collection. A gradient is the change in the value of a quantity, for example, concentration, pressure, or temperature with the change in another variable, usually distance. A change in c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
In Vitro
''In vitro'' (meaning in glass, or ''in the glass'') studies are performed with microorganisms, cells, or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in biology and its subdisciplines are traditionally done in labware such as test tubes, flasks, Petri dishes, and microtiter plates. Studies conducted using components of an organism that have been isolated from their usual biological surroundings permit a more detailed or more convenient analysis than can be done with whole organisms; however, results obtained from ''in vitro'' experiments may not fully or accurately predict the effects on a whole organism. In contrast to ''in vitro'' experiments, ''in vivo'' studies are those conducted in living organisms, including humans, and whole plants. Definition ''In vitro'' ( la, in glass; often not italicized in English usage) studies are conducted using components of an organism that have been isolated fro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Binding And Unbinding Mechanism Of LacI
Binding may refer to: Computing * Binding, associating a network socket with a local port number and IP address * Data binding, the technique of connecting two data elements together ** UI data binding, linking a user interface element to an element of a domain model, such as a database field ** XML data binding, representing XML document data using objects and classes * Key binding, or keyboard shortcut, mapping key combinations to software functionality * Language binding, a library providing a functional interface to second library in a different programming language * Name binding, the association of code or data with an identifier in a programming language ** Late binding, name binding which is resolved at run-time rather than in pre-execution time Science * Binding problem, a term for several problems in cognitive science and philosophy ** Neural binding, synchronous activity of neurons and neuronal ensembles * Molecular binding, an attractive interaction between two molecule ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
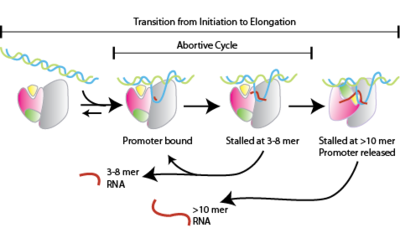
Abortive Initiation
Abortive initiation, also known as abortive transcription, is an early process of genetic transcription in which RNA polymerase binds to a DNA promoter and enters into cycles of synthesis of short mRNA transcripts which are released before the transcription complex leaves the promoter. This process occurs in both eukaryotes and prokaryotes. Abortive initiation is typically studied in the T3 and T7 RNA polymerases in bacteriophages and in ''E. coli''. Overall process Abortive initiation occurs prior to promoter clearance. # RNA polymerase binds to promoter DNA to form an RNA polymerase-promoter ''closed'' complex # RNA polymerase then unwinds one turn of DNA surrounding the transcription start site to yield an RNA polymerase-promoter ''open'' complex # RNA polymerase enters into abortive cycles of synthesis and releases short RNA products (contains less than 10 nucleotides) # RNA polymerase escapes the promoter and enters into the elongation step of transcription Mechanism Aborti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Motif
In a polymer, chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common Biomolecular structure#Tertiary structure, three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA. In nucleic acids Depending upon the sequence and other conditions, nucleic acids can form a variety of structural motifs which is thought to have biological significance. ;Stem-loop: Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tetrameric Protein
A tetrameric protein is a protein with a quaternary structure of four subunits (tetrameric). Homotetramers have four identical subunits (such as glutathione S-transferase), and heterotetramers are complexes of different subunits. A tetramer can be assembled as dimer of dimers with two homodimer subunits (such as sorbitol dehydrogenase), or two heterodimer subunits (such as hemoglobin). Subunit interactions in tetramers The interactions between subunits forming a tetramer is primarily determined by non covalent interaction. Hydrophobic effects, hydrogen bonds and electrostatic interactions are the primary sources for this binding process between subunits. For homotetrameric proteins such as Sorbitol dehydrogenase (SDH), the structure is believed to have evolved going from a monomeric to a dimeric and finally a tetrameric structure in evolution. The binding process in SDH and many other tetrameric enzymes can be described by the gain in free energy which can be determined from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |