|
LacUV5
The ''lacUV5'' promoter is a mutated promoter from the ''Escherichia coli'' lac operon which is used in molecular biology to drive gene expression on a plasmid. ''lacUV5'' is very similar to the classical ''lac'' promoter, containing just 2 base pair mutations in the -10 hexamer region, compared to the ''lac'' promoter. ''LacUV5'' is among the most commonly used promoters in molecular biology because it requires no additional activators and it drives high levels of gene expression. The ''lacUV5'' promoter sequence conforms more closely to the consensus sequence recognized by bacterial sigma factors than the traditional ''lac'' promoter does. Due to this, ''lacUV5'' recruits RNA Polymerase more effectively, thus leading to higher transcription of target genes. Additionally, unlike the ''lac'' promoter, ''lacUV5'' works independently of activator proteins or other cis regulatory elements (apart from the -10 and -35 promoter regions). While no activators are required, ''lacUV ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Activator (genetics)
A transcriptional activator is a protein (transcription factor) that increases transcription of a gene or set of genes. Activators are considered to have ''positive'' control over gene expression, as they function to promote gene transcription and, in some cases, are required for the transcription of genes to occur. Most activators are DNA-binding proteins that bind to enhancers or promoter-proximal elements. The DNA site bound by the activator is referred to as an "activator-binding site". The part of the activator that makes protein–protein interactions with the general transcription machinery is referred to as an "activating region" or "activation domain". Most activators function by binding sequence-specifically to a regulatory DNA site located near a promoter and making protein–protein interactions with the general transcription machinery (RNA polymerase and general transcription factors), thereby facilitating the binding of the general transcription machinery to the prom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
T7 RNA Polymerase
T7 RNA Polymerase is an RNA polymerase from the T7 bacteriophage that catalyzes the formation of RNA from DNA in the 5'→ 3' direction. Activity T7 polymerase is extremely promoter-specific and transcribes only DNA downstream of a T7 promoter. The T7 polymerase also requires a double stranded DNA template and Mg2+ ion as cofactor for the synthesis of RNA. It has a very low error rate. T7 polymerase has a molecular weight of 99 kDa. Promoter The promoter is recognized for binding and initiation of the transcription. The consensus in T7 and related phages is: 5' * 3' T7 TAATACGACTCACTATAGGGAGA T3 AATTAACCCTCACTAAAGGGAGA K11 AATTAGGGCACACTATAGGGAGA SP6 ATTTACGACACACTATAGAAGAA bind------------ -----------init Transcription begins at the asterisk-marked guanine. Structure T7 polymerase has been crystallised in several forms and the structures placed in the PDB. These explain how T7 polymerase binds to DNA and tra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone (i.e. each strand) of the DNA double helix. These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses. Inside a prokaryote, the restriction enzymes selectively cut up ''foreign'' DNA in a process called ''restriction digestion''; meanwhile, host DNA is protected by a modification enzyme (a methyltransferase) that modifi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HaeIII
''Hae''III is one of many restriction enzymes ( endonucleases) a type of prokaryotic DNA that protects organisms from unknown, foreign DNA. It is a restriction enzyme used in molecular biology laboratories. It was the third endonuclease to be isolated from the ''Haemophilus aegyptius'' bacteria. The enzyme's recognition site—the place where it cuts DNA molecules—is the GGCC nucleotide sequence which means it cleaves DNA at the site 5′-GG/CC-3. The recognition site is usually around 4-8 bps.This enzyme's gene has been sequenced and cloned. This is done to make DNA fragments in blunt ends. HaeIII is not effective for single stranded DNA cleavage. Properties ''Hae''III has a molecular weight of 37126. After a 2-10-fold of ''Hae''III takes place, there is overdigestion of a DNA substrate. This results in 100% being cut, more than 50% of fragments being ligated, and more than 95% being recut. Heat inactivation comes at about 80 °C for 20 minutes. The locus of the ''Hae ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
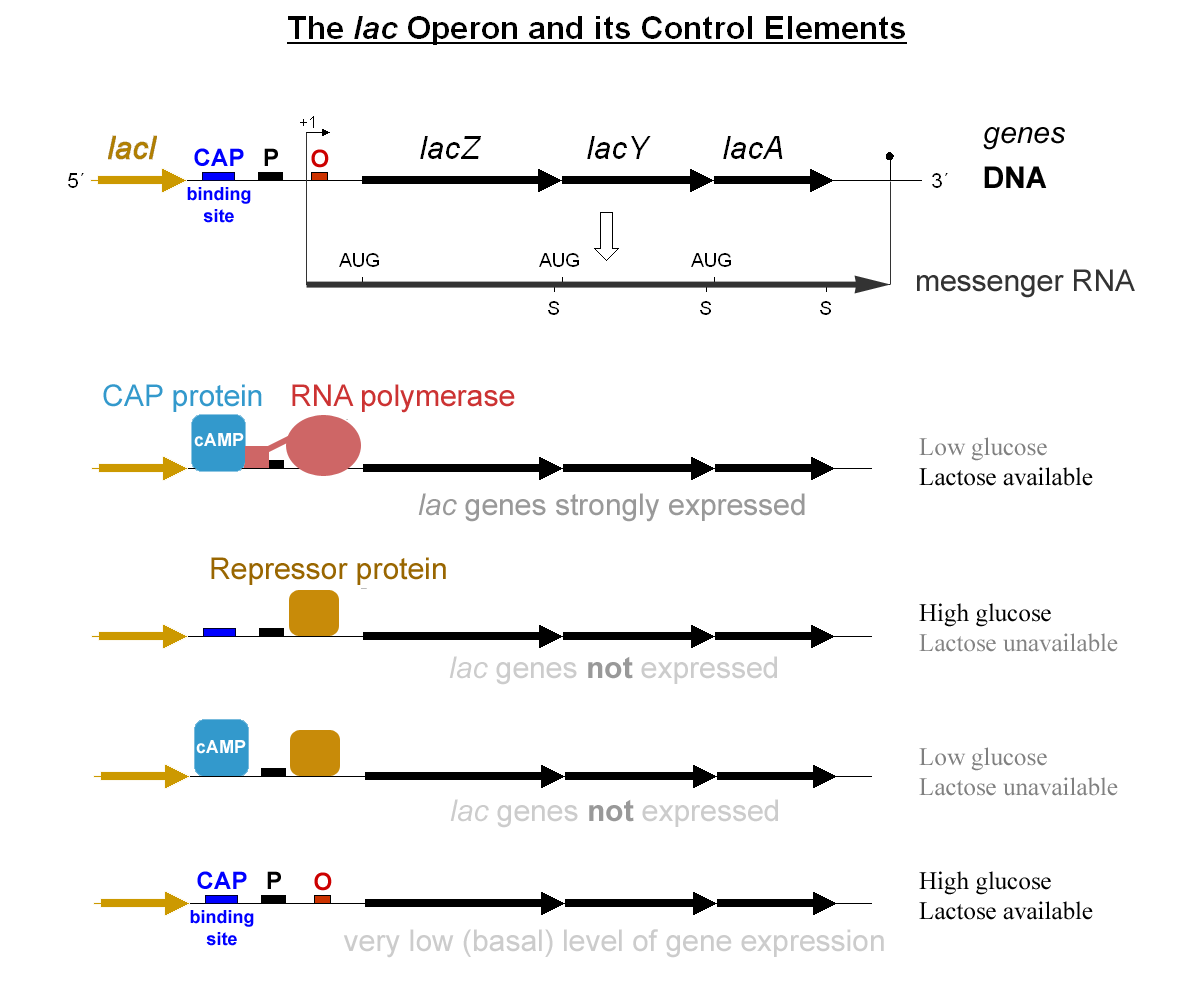
Lac Repressor
The ''lac'' repressor (LacI) is a DNA-binding protein that inhibits the expression of genes coding for proteins involved in the metabolism of lactose in bacteria. These genes are repressed when lactose is not available to the cell, ensuring that the bacterium only invests energy in the production of machinery necessary for uptake and utilization of lactose when lactose is present. When lactose becomes available, it is firstly converted into allolactose by β-Galactosidase (lacZ) in bacteria. The DNA binding ability of lac repressor bound with allolactose is inhibited due to allosteric regulation, thereby genes coding for proteins involved in lactose uptake and utilization can be expressed. Function The ''lac'' repressor (LacI) operates by a helix-turn-helix motif in its DNA-binding domain, binding base-specifically to the major groove of the operator region of the ''lac'' operon, with base contacts also made by residues of symmetry-related alpha helices, the "hinge" heli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory Element
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes ( pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides. RNAP produces RNA that, functionally, is either for protein coding, i.e. messenger RNA (mRNA); or n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Escherichia Coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escherichia'' that is commonly found in the lower intestine of warm-blooded organisms. Most ''E. coli'' strains are harmless, but some serotypes ( EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains do not cause disease in humans and are part of the normal microbiota of the gut; such strains are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of ''E. coli'' benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between ''E. col ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sigma Factor
A sigma factor (σ factor or specificity factor) is a protein needed for initiation of transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to gene promoters. It is homologous to archaeal transcription factor B and to eukaryotic factor TFIIB. The specific sigma factor used to initiate transcription of a given gene will vary, depending on the gene and on the environmental signals needed to initiate transcription of that gene. Selection of promoters by RNA polymerase is dependent on the sigma factor that associates with it. They are also found in plant chloroplasts as a part of the bacteria-like plastid-encoded polymerase (PEP). The sigma factor, together with RNA polymerase, is known as the RNA polymerase holoenzyme. Every molecule of RNA polymerase holoenzyme contains exactly one sigma factor subunit, which in the model bacterium ''Escherichia coli'' is one of those listed below. The number of sigma f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Consensus Sequence
In molecular biology and bioinformatics, the consensus sequence (or canonical sequence) is the calculated order of most frequent residues, either nucleotide or amino acid, found at each position in a sequence alignment. It serves as a simplified representation of the population. It represents the results of multiple sequence alignments in which related sequences are compared to each other and similar sequence motifs are calculated. Such information is important when considering sequence-dependent enzymes such as RNA polymerase.Pierce, Benjamin A. 2002. Genetics : A Conceptual Approach. 1st ed. New York: W.H. Freeman and Co. Biological significance A protein binding site, represented by a consensus sequence, may be a short sequence of nucleotides which is found several times in the genome and is thought to play the same role in its different locations. For example, many transcription factors recognize particular patterns in the promoters of the genes they regulate. In the same way, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |