|
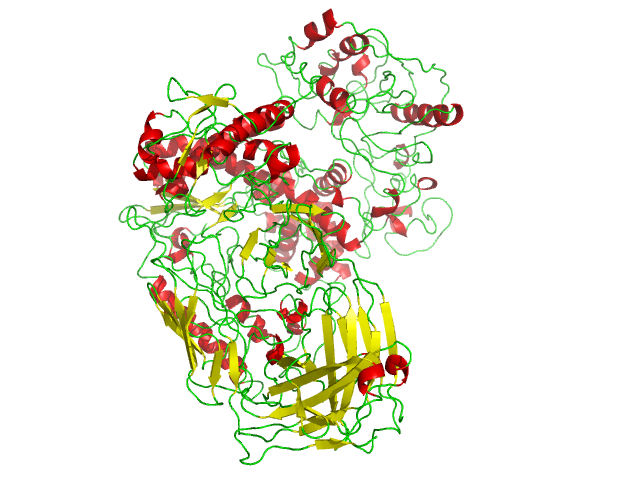
Klenow Fragment
The Klenow fragment is a large protein fragment produced when DNA polymerase I from '' E. coli'' is enzymatically cleaved by the protease subtilisin. First reported in 1970, it retains the 5' → 3' polymerase activity and the 3’ → 5’ exonuclease activity for removal of precoding nucleotides and proofreading, but loses its 5' → 3' exonuclease activity. The other smaller fragment formed when DNA polymerase I from '' E. coli'' is cleaved by subtilisin retains the 5' → 3' exonuclease activity but does not have the other two activities exhibited by the Klenow fragment (i.e. 5' → 3' polymerase activity, and 3' → 5' exonuclease activity). Research Because the 5' → 3' exonuclease activity of DNA polymerase I from '' E. coli'' makes it unsuitable for many applications, the Klenow fragment, which lacks this activity, can be very useful in research. The Klenow fragment is extremely useful for research-based tasks such as: * Synthesis of double-stranded DNA from single-s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Polymerase I
DNA polymerase I (or Pol I) is an enzyme that participates in the process of prokaryotic DNA replication. Discovered by Arthur Kornberg in 1956, it was the first known DNA polymerase (and the first known of any kind of polymerase). It was initially characterized in '' E. coli'' and is ubiquitous in prokaryotes. In ''E. coli'' and many other bacteria, the gene that encodes Pol I is known as ''polA''. The ''E. coli'' Pol I enzyme is composed of 928 amino acids, and is an example of a processive enzyme — it can sequentially catalyze multiple polymerisation steps without releasing the single-stranded template. The physiological function of Pol I is mainly to support repair of damaged DNA, but it also contributes to connecting Okazaki fragments by deleting RNA primers and replacing the ribonucleotides with DNA. Discovery In 1956, Arthur Kornberg and colleagues discovered Pol I by using ''Escherichia coli'' (''E. coli'') extracts to develop a DNA synthesis assay. The scientist ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Hierarchy of proteases Based on catalytic residue Proteases can be classified into seven broad groups: * Serine protease ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Subtilisin
Subtilisin is a protease (a protein-digesting enzyme) initially obtained from ''Bacillus subtilis''. Subtilisins belong to subtilases, a group of serine proteases that – like all serine proteases – initiate the nucleophilic attack on the peptide (amide) bond through a serine residue at the active site. Subtilisins typically have molecular weights 27kDa. They can be obtained from certain types of soil bacteria, for example, ''Bacillus amyloliquefaciens'' from which they are secreted in large amounts. Nomenclature Subtilisin is also commercially known as ''Alcalase®'', ''Endocut-02L'', ''ALK-enzyme'', ''bacillopeptidase'', ''Bacillus subtilis alkaline proteinase bioprase'', ''bioprase AL'', ''colistinase'', ''genenase I'', ''Esperase®'', ''maxatase'', ''protease XXVII'', ''thermoase'', ''superase'', ''subtilisin DY'', ''subtilopeptidase'', ''SP 266'', ''Savinase®'', ''kazusase'', ''protease VIII'', ''protin A 3L'', ''Savinase®'', ''orientase 10B'', ''protease S.'' It ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase
A polymerase is an enzyme ( EC 2.7.7.6/7/19/48/49) that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base-pairing interactions or RNA by half ladder replication. A DNA polymerase from the thermophilic bacterium, ''Thermus aquaticus'' (''Taq'') ( PDBbr>1BGX EC 2.7.7.7) is used in the polymerase chain reaction, an important technique of molecular biology. A polymerase may be template dependent or template independent. Poly-A-polymerase is an example of template independent polymerase. Terminal deoxynucleotidyl transferase also known to have template independent and template dependent activities. Types By function *DNA polymerase (DNA-directed DNA polymerase, DdDP) **Family A: DNA polymerase I; Pol γ, θ, ν **Family B: DNA polymerase II; Pol α, δ, ε, ζ **Family C: DNA polymerase III holoenzyme **Family X: Pol β, λ, μ * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease. In both archaea and eukaryotes, one of the main routes of RNA degradation is performed by the multi-protein exosome complex, which consists largely of 3′ to 5′ exoribonucleases. Significance to polymerase RNA polymerase II is known to be in effect during transcriptional termination; it works with a 5' exonuclease (human gene Xrn2) to degrade the newly formed transcript ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Radioactive
Radioactive decay (also known as nuclear decay, radioactivity, radioactive disintegration, or nuclear disintegration) is the process by which an unstable atomic nucleus loses energy by radiation. A material containing unstable nuclei is considered radioactive. Three of the most common types of decay are alpha decay ( ), beta decay ( ), and gamma decay ( ), all of which involve emitting one or more particles. The weak force is the mechanism that is responsible for beta decay, while the other two are governed by the electromagnetism and nuclear force. A fourth type of common decay is electron capture, in which an unstable nucleus captures an inner electron from one of the electron shells. The loss of that electron from the shell results in a cascade of electrons dropping down to that lower shell resulting in emission of discrete X-rays from the transitions. A common example is iodine-125 commonly used in medical settings. Radioactive decay is a stochastic (i.e. random) process ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Probe
In molecular biology, a hybridization probe (HP) is a fragment of DNA or RNA of usually 15–10000 nucleotide long which can be radioactively or fluorescently labeled. HP can be used to detect the presence of nucleotide sequences in analyzed RNA or DNA that are complementary to the sequence in the probe. The labeled probe is first denatured (by heating or under alkaline conditions such as exposure to sodium hydroxide) into single stranded DNA (ssDNA) and then hybridized to the target ssDNA ( Southern blotting) or RNA (northern blotting) immobilized on a membrane or in situ. To detect hybridization of the probe to its target sequence, the probe is tagged (or "labeled") with a molecular marker of either radioactive or (more recently) fluorescent molecules. Commonly used markers are 32P (a radioactive isotope of phosphorus incorporated into the phosphodiester bond in the probe DNA), digoxigenin, a non-radioactive, antibody-based marker, biotin or fluorescein. DNA sequences o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Taq Polymerase
''Taq'' polymerase is a thermostable DNA polymerase I named after the thermophilic eubacterial microorganism ''Thermus aquaticus,'' from which it was originally isolated by Chien et al. in 1976. Its name is often abbreviated to ''Taq'' or ''Taq'' pol. It is frequently used in the polymerase chain reaction (PCR), a method for greatly amplifying the quantity of short segments of DNA. ''T. aquaticus'' is a bacterium that lives in hot springs and hydrothermal vents, and ''Taq'' polymerase was identified as an enzyme able to withstand the protein-denaturing conditions (high temperature) required during PCR. Therefore, it replaced the DNA polymerase from '' E. coli'' originally used in PCR. Enzymatic properties ''Taqs optimum temperature for activity is 75–80 °C, with a half-life of greater than 2 hours at 92.5 °C, 40 minutes at 95 °C and 9 minutes at 97.5 °C, and can replicate a 1000 base pair strand of DNA in less than 10 seconds at 72 °C. At ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |