|
Importin
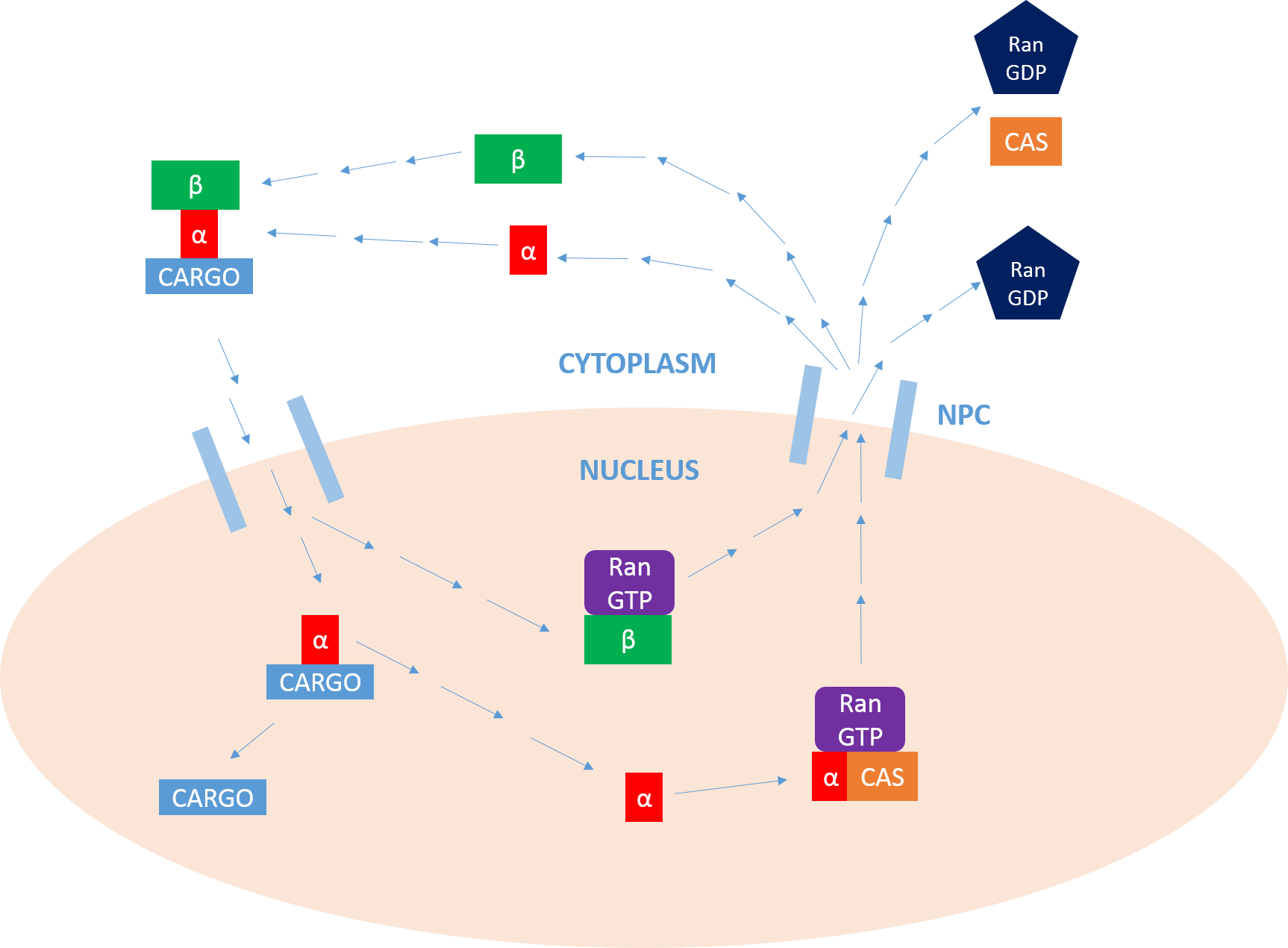
Importin is a type of karyopherin that transports protein molecules from the cell's cytoplasm to the nucleus. It does so by binding to specific recognition sequences, called nuclear localization sequences (NLS). Importin has two subunits, importin α and importin β. Members of the importin-β family can bind and transport cargo by themselves, or can form heterodimers with importin-α. As part of a heterodimer, importin-β mediates interactions with the pore complex, while importin-α acts as an adaptor protein to bind the nuclear localization signal (NLS) on the cargo. The NLS-Importin α-Importin β trimer dissociates after binding to Ran GTP inside the nucleus, with the two importin proteins being recycled to the cytoplasm for further use. Discovery Importin can exist as either a heterodimer of importin-α/β or as a monomer of Importin-β. Importin-α was first isolated in 1994 by a group includinEnno Hartmann based at the Max Delbrück Center for Molecular Medicine. The p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Pore
A nuclear pore is a part of a large complex of proteins, known as a nuclear pore complex that spans the nuclear envelope, which is the double membrane surrounding the eukaryotic cell nucleus. There are approximately 1,000 nuclear pore complexes (NPCs) in the nuclear envelope of a vertebrate cell, but this number varies depending on cell type and the stage in the life cycle. The human nuclear pore complex (hNPC) is a 110 megadalton (MDa) structure. The proteins that make up the nuclear pore complex are known as nucleoporins; each NPC contains at least 456 individual protein molecules and is composed of 34 distinct nucleoporin proteins. About half of the nucleoporins typically contain solenoid protein domains—either an alpha solenoid or a beta-propeller fold, or in some cases both as separate structural domains. The other half show structural characteristics typical of "natively unfolded" or intrinsically disordered proteins, i.e. they are highly flexible proteins that lack ord ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Localization Sequence
A nuclear localization signal ''or'' sequence (NLS) is an amino acid sequence that 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines or arginines exposed on the protein surface. Different nuclear localized proteins may share the same NLS. An NLS has the opposite function of a nuclear export signal (NES), which targets proteins out of the nucleus. Types Classical These types of NLSs can be further classified as either monopartite or bipartite. The major structural differences between the two are that the two basic amino acid clusters in bipartite NLSs are separated by a relatively short spacer sequence (hence bipartite - 2 parts), while monopartite NLSs are not. The first NLS to be discovered was the sequence PKKKRKV in the SV40 Large T-antigen (a monopartite NLS). The NLS of nucleoplasmin, KR AATKKAGQAKKK, is the prototype of the ubiquitous bipartite signal: two cluster ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Localization Signal
A nuclear localization signal ''or'' sequence (NLS) is an amino acid sequence that 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines or arginines exposed on the protein surface. Different nuclear localized proteins may share the same NLS. An NLS has the opposite function of a nuclear export signal (NES), which targets proteins out of the nucleus. Types Classical These types of NLSs can be further classified as either monopartite or bipartite. The major structural differences between the two are that the two basic amino acid clusters in bipartite NLSs are separated by a relatively short spacer sequence (hence bipartite - 2 parts), while monopartite NLSs are not. The first NLS to be discovered was the sequence PKKKRKV in the SV40 Large T-antigen (a monopartite NLS). The NLS of nucleoplasmin, KR AATKKAGQAKKK, is the prototype of the ubiquitous bipartite signal: two cluster ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Karyopherin
Karyopherins are proteins involved in transporting molecules between the cytoplasm and the nucleus of a eukaryotic cell. The inside of the nucleus is called the karyoplasm (or nucleoplasm). Generally, karyopherin-mediated transport occurs through nuclear pores which acts as a gateway into and out of the nucleus. Most proteins require karyopherins to traverse the nuclear pore. Karyopherins can act as ''importins'' (i.e. helping proteins get into the nucleus) or ''exportins'' (i.e. helping proteins get out of the nucleus). They belong to the nuclear pore complex family in the transporter classification database (TCDB). Energy for transport is derived from the Ran gradient. Upon stress, several karyopherins stop shuttling between the nucleus and the cytoplasm and are sequestered in stress granules, cytoplasmic aggregates of ribonucleoprotein complexes. Importin beta Importin beta is a variety of karyopherin that facilitates the transport of cargo proteins into the nucleus. First, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
KPNB1
Importin subunit beta-1 is a protein that in humans is encoded by the ''KPNB1'' gene. Function Nucleocytoplasmic transport, a signal- and energy-dependent process, takes place through nuclear pore complexes embedded in the nuclear envelope. The import of proteins containing a classical nuclear localization signal (NLS) requires the NLS import receptor, a heterodimer of importin alpha and beta subunits. Each of these subunits are part of the karyopherin family of proteins. Importin alpha binds the NLS-containing cargo in the cytoplasm and importin beta docks the complex at the cytoplasmic side of the nuclear pore complex. In the presence of nucleoside triphosphates and the small GTP binding protein Ran, the complex moves into the nuclear pore complex and the importin subunits dissociate. Importin alpha enters the nucleoplasm with its passenger protein and importin beta remains at the pore. Interactions between importin beta and the FG repeats of nucleoporins are essential in tra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
IPO4
Importin-4 is a protein that in humans is encoded by the ''IPO4'' gene In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b .... References Further reading * * * * * * * {{gene-14-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
KPNA1
Importin subunit alpha-5 is a protein that in humans is encoded by the ''KPNA1'' gene. Interactions Importin subunit alpha-5 has been shown to interact with KPNB1 and UBR5 E3 ubiquitin-protein ligase UBR5 is an enzyme that in humans is encoded by the ''UBR5'' gene. Function This gene encodes a progestin-induced protein, which belongs to the HECT (homology to E6-AP carboxyl terminus) family. The HECT family protei .... References Further reading * * * * * * * * * * * * * * * * * * * Armadillo-repeat-containing proteins {{gene-3-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
IPO7
Importin-7 is a protein that in humans is encoded by the ''IPO7'' gene. The importin-alpha/beta complex and the GTPase Ran mediate nuclear import of proteins with a classical nuclear localization signal. The protein encoded by this gene is a member of a class of approximately 20 potential Ran targets that share a sequence motif related to the Ran-binding site of importin-beta. Similar to importin-beta, this protein prevents the activation of Ran's GTPase by RanGAP1 and inhibits nucleotide exchange on RanGTP, and also binds directly to nuclear pore complexes where it competes for binding sites with importin-beta and transportin. This protein has a Ran-dependent transport cycle and it can cross the nuclear envelope rapidly and in both directions. At least four importin beta-like transport receptors, namely importin beta itself, transportin, RanBP5 and RanBP7, directly bind and import ribosomal proteins A ribosomal protein (r-protein or rProtein) is any of the proteins that, in co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology (biology)
In biology, homology is similarity due to shared ancestry between a pair of structures or genes in different taxa. A common example of homologous structures is the forelimbs of vertebrates, where the wings of bats and birds, the arms of primates, the front flippers of whales and the forelegs of four-legged vertebrates like dogs and crocodiles are all derived from the same ancestral tetrapod structure. Evolutionary biology explains homologous structures adapted to different purposes as the result of descent with modification from a common ancestor. The term was first applied to biology in a non-evolutionary context by the anatomist Richard Owen in 1843. Homology was later explained by Charles Darwin's theory of evolution in 1859, but had been observed before this, from Aristotle onwards, and it was explicitly analysed by Pierre Belon in 1555. In developmental biology, organs that developed in the embryo in the same manner and from similar origins, such as from matching p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signal Transducing Adaptor Protein
Signal transducing adaptor proteins (STAPs) are proteins that are accessory to main proteins in a signal transduction pathway. Adaptor proteins contain a variety of protein-binding modules that link protein-binding partners together and facilitate the creation of larger signaling complexes. These proteins tend to lack any intrinsic enzymatic activity themselves, instead mediating specific protein–protein interactions that drive the formation of protein complexes. Examples of adaptor proteins include MYD88, Grb2 and SHC1. Signaling components Much of the specificity of signal transduction depends on the recruitment of several signalling components such as protein kinases and G-protein GTPases into short-lived active complexes in response to an activating signal such as a growth factor binding to its receptor. Domains Adaptor proteins usually contain several domains within their structure (e.g., Src homology 2 (SH2) and SH3 domains) that allow specific interactions with sev ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences, such as calculating the distance cost between strings in a natural language or in financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence alignments of proteins, the degree of similarity between amino acids occupying a parti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Armadillo Repeat
An armadillo repeat is the name of a characteristic, repetitive amino acid sequence of about 40 residues in length that is found in many proteins. Proteins that contain armadillo repeats typically contain several tandemly repeated copies. Each armadillo repeat is composed of a pair of alpha helices that form a hairpin structure. Multiple copies of the repeat form what is known as an alpha solenoid structure. Examples of proteins that contain armadillo repeats include β-catenin, α-importin, plakoglobin, adenomatous polyposis coli (APC), and many others. The term armadillo derives from the historical name of the β-catenin gene in the fruitfly ''Drosophila'' where the armadillo repeat was first discovered. Although β-catenin was previously believed to be a protein involved in linking cadherin cell adhesion proteins to the cytoskeleton, recent work indicates that β-catenin regulates the homodimerization of alpha-catenin, which in turn controls actin branching and bundling.Nus ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |