|
Isomorph (gene)
After Muller's classification was described as an isomorph, or gene mutation that expresses a nonsense point mutant with expression identical to the original allele. Therefore, in respect of the relationships between the original and mutated genes it is difficult to ascertain the effects of dominanceness or/and recessiveness. ;Muller’s classification of mutant alleles See also *Allele *Mutation *Muller's morphs Hermann J. Muller (1890–1967), who was a 1946 Nobel Prize winner, coined the terms amorph, hypomorph, hypermorph, antimorph and neomorph to classify mutations based on their behaviour in various genetic situations, as well as gene interac ... References {{reflist Mutation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA (such as pyrimidine dimers caused by exposure to ultraviolet radiation), which then may undergo error-prone repair (especially microhomology-mediated end joining), cause an error during other forms of repair, or cause an error during replication (translesion synthesis). Mutations may also result from insertion or deletion of segments of DNA due to mobile genetic elements. Mutations may or may not produce detectable changes in the observable characteristics (phenotype) of an organism. Mutations play a part in both normal and abnormal biological processes including: evolution, cancer, and the development of the immune system, including junctional diversity. Mutation is the ultimate source of a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
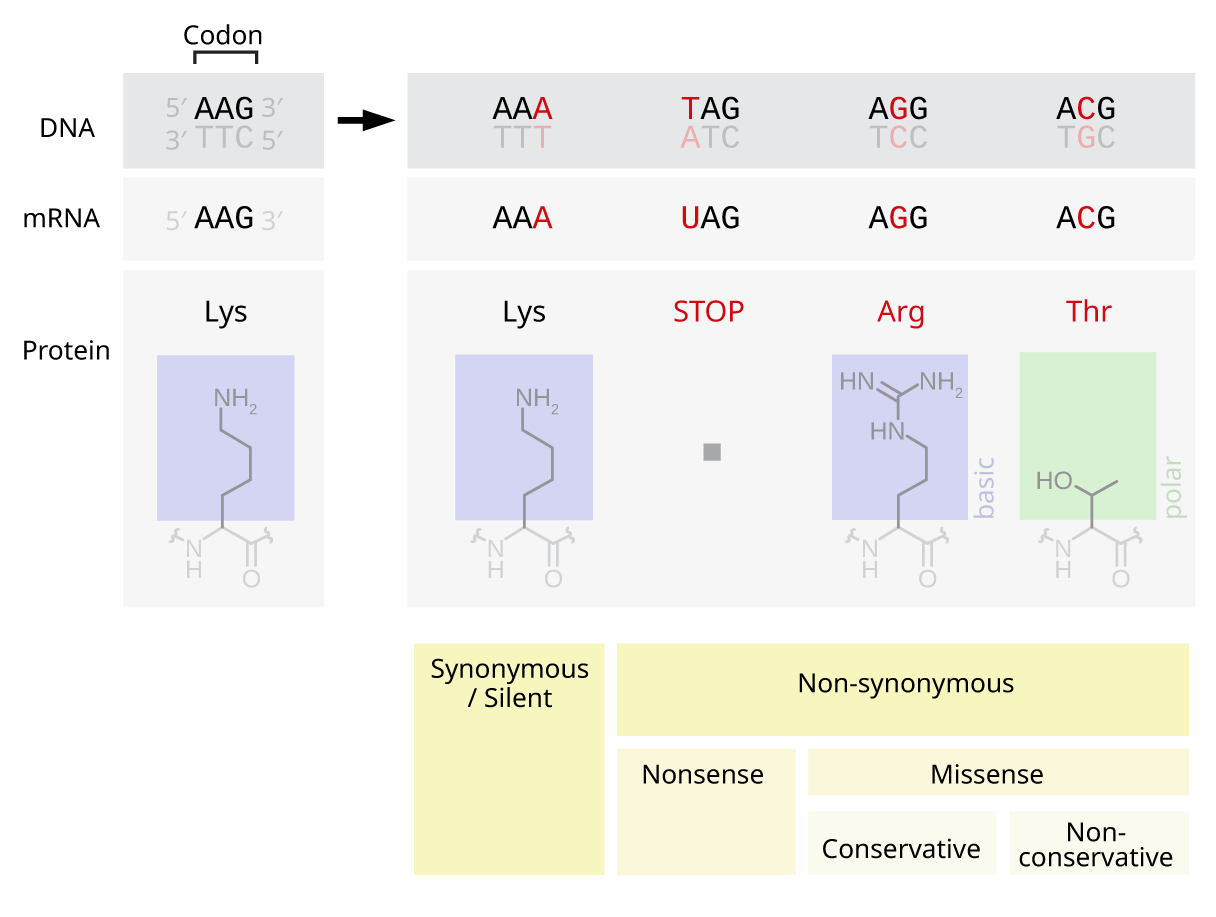
Nonsense Mutation
In genetics, a nonsense mutation is a point mutation in a sequence of DNA that results in a premature stop codon, or a ''nonsense codon'' in the transcribed mRNA, and in leading to a truncated, incomplete, and usually nonfunctional protein product. The functional effect of a nonsense mutation depends on the location of the stop codon within the coding DNA. For example, the effect of a nonsense mutation depends on the proximity of the nonsense mutation to the original stop codon, and the degree to which functional subdomains of the protein are affected. As nonsense mutations leads to premature termination of polypeptide chains; they are also called chain termination mutations. Missense mutations differ from nonsense mutations since they are point mutations that exhibit a single nucleotide change to cause substitution of a different amino acid. A nonsense mutation also differs from a nonstop mutation, which is a point mutation that removes a stop codon. About 10% of patients facin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Point Mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences that are moderately predictable based upon the specifics of the mutation. These consequences can range from no effect (e.g. synonymous mutations) to deleterious effects (e.g. frameshift mutations), with regard to protein production, composition, and function. Causes Point mutations usually take place during DNA replication. DNA replication occurs when one double-stranded DNA molecule creates two single strands of DNA, each of which is a template for the creation of the complementary strand. A single point mutation can change the whole DNA sequence. Changing one purine or pyrimidine may change the amino acid that the nucleotides code for. Point mutations may arise from spontaneous mutations that occur during DNA replication. The rate of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Allele
An allele (, ; ; modern formation from Greek ἄλλος ''állos'', "other") is a variation of the same sequence of nucleotides at the same place on a long DNA molecule, as described in leading textbooks on genetics and evolution. ::"The chromosomal or genomic location of a gene or any other genetic element is called a locus (plural: loci) and alternative DNA sequences at a locus are called alleles." The simplest alleles are single nucleotide polymorphisms (SNP). but they can also be insertions and deletions of up to several thousand base pairs. Popular definitions of 'allele' typically refer only to different alleles within genes. For example, the ABO blood grouping is controlled by the ABO gene, which has six common alleles (variants). In population genetics, nearly every living human's phenotype for the ABO gene is some combination of just these six alleles. Most alleles observed result in little or no change in the function of the gene product it codes for. However, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dominance (genetics)
In genetics, dominance is the phenomenon of one variant (allele) of a gene on a chromosome masking or overriding the effect of a different variant of the same gene on the other copy of the chromosome. The first variant is termed dominant and the second recessive. This state of having two different variants of the same gene on each chromosome is originally caused by a mutation in one of the genes, either new (''de novo'') or inherited. The terms autosomal dominant or autosomal recessive are used to describe gene variants on non-sex chromosomes ( autosomes) and their associated traits, while those on sex chromosomes (allosomes) are termed X-linked dominant, X-linked recessive or Y-linked; these have an inheritance and presentation pattern that depends on the sex of both the parent and the child (see Sex linkage). Since there is only one copy of the Y chromosome, Y-linked traits cannot be dominant or recessive. Additionally, there are other forms of dominance such as incomplete d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Recessiveness
In genetics, dominance is the phenomenon of one variant (allele) of a gene on a chromosome masking or overriding the effect of a different variant of the same gene on the other copy of the chromosome. The first variant is termed dominant and the second recessive. This state of having two different variants of the same gene on each chromosome is originally caused by a mutation in one of the genes, either new (''de novo'') or inherited. The terms autosomal dominant or autosomal recessive are used to describe gene variants on non-sex chromosomes ( autosomes) and their associated traits, while those on sex chromosomes (allosomes) are termed X-linked dominant, X-linked recessive or Y-linked; these have an inheritance and presentation pattern that depends on the sex of both the parent and the child (see Sex linkage). Since there is only one copy of the Y chromosome, Y-linked traits cannot be dominant or recessive. Additionally, there are other forms of dominance such as incomplete d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Wild Type
The wild type (WT) is the phenotype of the typical form of a species as it occurs in nature. Originally, the wild type was conceptualized as a product of the standard "normal" allele at a locus, in contrast to that produced by a non-standard, "mutant" allele. "Mutant" alleles can vary to a great extent, and even become the wild type if a genetic shift occurs within the population. Continued advancements in genetic mapping technologies have created a better understanding of how mutations occur and interact with other genes to alter phenotype. It is now appreciated that most or all gene loci exist in a variety of allelic forms, which vary in frequency throughout the geographic range of a species, and that a uniform wild type does not exist. In general, however, the most prevalent allele – i.e., the one with the highest gene frequency – is the one deemed wild type. The concept of wild type is useful in some experimental organisms such as fruit flies ''Drosophila melanogaster'' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Muller's Morphs
Hermann J. Muller (1890–1967), who was a 1946 Nobel Prize winner, coined the terms amorph, hypomorph, hypermorph, antimorph and neomorph to classify mutations based on their behaviour in various genetic situations, as well as gene interaction between themselves.Muller, H. J. 1932. Further studies on the nature and causes of gene mutations. ''Proceedings of the 6th International Congress of Genetics'', pp. 213–255. These classifications are still widely used in ''Drosophila'' genetics to describe mutations. For a more general description of mutations, see mutation, and for a discussion of allele interactions, see dominance relationship. ''Key: In the following sections, alleles are referred to as +=wildtype, m=mutant, Df=gene deletion, Dp=gene duplication. Phenotypes are compared with '>', meaning 'phenotype is more severe than Loss of function Amorph Amorphic describes a mutation that causes complete loss of gene function. Amorph is sometimes used interchangeably ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hypomorph
Hermann J. Muller (1890–1967), who was a 1946 Nobel Prize winner, coined the terms amorph, hypomorph, hypermorph, antimorph and neomorph to classify mutations based on their behaviour in various genetic situations, as well as gene interaction between themselves.Muller, H. J. 1932. Further studies on the nature and causes of gene mutations. ''Proceedings of the 6th International Congress of Genetics'', pp. 213–255. These classifications are still widely used in ''Drosophila'' genetics to describe mutations. For a more general description of mutations, see mutation, and for a discussion of allele interactions, see dominance relationship. ''Key: In the following sections, alleles are referred to as +=wildtype, m=mutant, Df=gene deletion, Dp=gene duplication. Phenotypes are compared with '>', meaning 'phenotype is more severe than Loss of function Amorph Amorphic describes a mutation that causes complete loss of gene function. Amorph is sometimes used interchangeably w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hypermorph
Hermann J. Muller (1890–1967), who was a 1946 Nobel Prize winner, coined the terms amorph, hypomorph, hypermorph, antimorph and neomorph to classify mutations based on their behaviour in various genetic situations, as well as gene interaction between themselves.Muller, H. J. 1932. Further studies on the nature and causes of gene mutations. ''Proceedings of the 6th International Congress of Genetics'', pp. 213–255. These classifications are still widely used in ''Drosophila'' genetics to describe mutations. For a more general description of mutations, see mutation, and for a discussion of allele interactions, see dominance relationship. ''Key: In the following sections, alleles are referred to as +=wildtype, m=mutant, Df=gene deletion, Dp=gene duplication. Phenotypes are compared with '>', meaning 'phenotype is more severe than Loss of function Amorph Amorphic describes a mutation that causes complete loss of gene function. Amorph is sometimes used interchangeably w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neomorph
Hermann J. Muller (1890–1967), who was a 1946 Nobel Prize winner, coined the terms amorph, hypomorph, hypermorph, antimorph and neomorph to classify mutations based on their behaviour in various genetic situations, as well as gene interaction between themselves.Muller, H. J. 1932. Further studies on the nature and causes of gene mutations. ''Proceedings of the 6th International Congress of Genetics'', pp. 213–255. These classifications are still widely used in ''Drosophila'' genetics to describe mutations. For a more general description of mutations, see mutation, and for a discussion of allele interactions, see dominance relationship. ''Key: In the following sections, alleles are referred to as +=wildtype, m=mutant, Df=gene deletion, Dp=gene duplication. Phenotypes are compared with '>', meaning 'phenotype is more severe than Loss of function Amorph Amorphic describes a mutation that causes complete loss of gene function. Amorph is sometimes used interchangeably w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |



_(14582377398).jpg)