|
In Silico
In biology and other experimental sciences, an ''in silico'' experiment is one performed on computer or via computer simulation. The phrase is pseudo-Latin for 'in silicon' (correct la, in silicio), referring to silicon in computer chips. It was coined in 1987 as an allusion to the Latin phrases , , and , which are commonly used in biology Biology is the scientific study of life. It is a natural science with a broad scope but has several unifying themes that tie it together as a single, coherent field. For instance, all organisms are made up of cells that process hereditar ... (especially systems biology). The latter phrases refer, respectively, to experiments done in living organisms, outside living organisms, and where they are found in nature. History The earliest known use of the phrase was by Christopher Langton to describe artificial life, in the announcement of a workshop on that subject at the Center for Nonlinear Studies at the Los Alamos National Laborat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Forest Of Synthetic Pyramidal Dendrites Grown Using Cajal's Laws Of Neuronal Branching
A forest is an area of land dominated by trees. Hundreds of definitions of forest are used throughout the world, incorporating factors such as tree density, tree height, land use, legal standing, and ecological function. The United Nations' Food and Agriculture Organization (FAO) defines a forest as, "Land spanning more than 0.5 hectares with trees higher than 5 meters and a Canopy (biology), canopy cover of more than 10 percent, or trees able to reach these thresholds ''in situ''. It does not include land that is predominantly under agricultural or urban use." Using this definition, ''Global Forest Resources Assessment (FRA), Global Forest Resources Assessment 2020'' (FRA 2020) found that forests covered , or approximately 31 percent of the world's land area in 2020. Forests are the predominant terrestrial ecosystem of Earth, and are found around the globe. More than half of the world's forests are found in only five countries (Brazil, Canada, China, Russia, and the United S ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drug Discovery
In the fields of medicine, biotechnology and pharmacology, drug discovery is the process by which new candidate medications are discovered. Historically, drugs were discovered by identifying the active ingredient from traditional remedies or by serendipitous discovery, as with penicillin. More recently, chemical libraries of synthetic small molecules, natural products or extracts were screened in intact cells or whole organisms to identify substances that had a desirable therapeutic effect in a process known as classical pharmacology. After sequencing of the human genome allowed rapid cloning and synthesis of large quantities of purified proteins, it has become common practice to use high throughput screening of large compounds libraries against isolated biological targets which are hypothesized to be disease-modifying in a process known as reverse pharmacology. Hits from these screens are then tested in cells and then in animals for efficacy. Modern drug discovery i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotic
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacteria and Archaea (both prokaryotes) make up the other two domains. The eukaryotes are usually now regarded as having emerged in the Archaea or as a sister of the Asgard archaea. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among archaea. Eukaryotes represent a small minority of the number of organisms, but, due to their generally much larger size, their collective global biomass is estimated to be about equal to that of prokaryotes. Eukaryotes emerged approximately 2.3–1.8 billion years ago, during the Proterozoic eon, likely as flagellated phagotrophs. Their name comes from the Greek εὖ (''eu'', "well" or "good") and κάρυον (''karyon'', "nut" or "kernel"). E ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Prokaryotic
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Connections". Pearson Education. San Francisco: 2003. In the two-empire system arising from the work of Édouard Chatton, prokaryotes were classified within the empire Prokaryota. But in the three-domain system, based upon molecular analysis, prokaryotes are divided into two domains: ''Bacteria'' (formerly Eubacteria) and ''Archaea'' (formerly Archaebacteria). Organisms with nuclei are placed in a third domain, Eukaryota. In the study of the origins of life, prokaryotes are thought to have arisen before eukaryotes. Besides the absence of a nucleus, prokaryotes also lack mitochondria, or most of the other membrane-bound organelles that characterize the eukaryotic cell. It was once thought that prokaryotic cellular components within the cytopla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Artificial Gene Synthesis
Artificial gene synthesis, or simply gene synthesis, refers to a group of methods that are used in synthetic biology to construct and assemble genes from nucleotides '' de novo''. Unlike DNA synthesis in living cells, artificial gene synthesis does not require template DNA, allowing virtually any DNA sequence to be synthesized in the laboratory. It comprises two main steps, the first of which is solid-phase DNA synthesis, sometimes known as DNA printing. This produces oligonucleotide fragments that are generally under 200 base pairs. The second step then involves connecting these oligonucleotide fragments using various DNA assembly methods. Because artificial gene synthesis does not require template DNA, it is theoretically possible to make a completely synthetic DNA molecule with no limits on the nucleotide sequence or size. Synthesis of the first complete gene, a yeast tRNA, was demonstrated by Har Gobind Khorana and coworkers in 1972. Synthesis of the first peptide- and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Analysis
In bioinformatics, sequence analysis is the process of subjecting a DNA, RNA or peptide sequence to any of a wide range of analytical methods to understand its features, function, structure, or evolution. Methodologies used include sequence alignment, searches against biological databases, and others. Since the development of methods of high-throughput production of gene and protein sequences, the rate of addition of new sequences to the databases increased very rapidly. Such a collection of sequences does not, by itself, increase the scientist's understanding of the biology of organisms. However, comparing these new sequences to those with known functions is a key way of understanding the biology of an organism from which the new sequence comes. Thus, sequence analysis can be used to assign function to genes and proteins by the study of the similarities between the compared sequences. Nowadays, there are many tools and techniques that provide the sequence comparisons (sequence a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Database
In the field of bioinformatics, a sequence database is a type of biological database that is composed of a large collection of computerized ("digital") nucleic acid sequences, protein sequences, or other polymer sequences stored on a computer. The UniProt database is an example of a protein sequence database. As of 2013 it contained over 40 million sequences and is growing at an exponential rate. Historically, sequences were published in paper form, but as the number of sequences grew, this storage method became unsustainable. Search Searching in a sequence database involves looking for similarities between a genomic/protein sequence and a query string and, finding the sequence in the database that "best" matches the target sequence (based on criteria which vary depending on the search method). The number of matches/hits is used to formulate a score that determines the similarity between the sequence query and the sequences in the sequence database. The main goal is to have a goo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Sequence
A nucleic acid sequence is a succession of Nucleobase, bases signified by a series of a set of five different letters that indicate the order of nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule. By convention, sequences are usually presented from the Directionality (molecular biology), 5' end to the 3' end. For DNA, the Sense (molecular biology), sense strand is used. Because nucleic acids are normally linear (unbranched) polymers, specifying the sequence is equivalent to defining the covalent structure of the entire molecule. For this reason, the nucleic acid sequence is also termed the Biomolecular structure#Primary structure, primary structure. The sequence has capacity to represent information. Biological deoxyribonucleic acid represents the information which directs the functions of an organism. Nucleic acids also have a Nucleic acid secondary structure, secondary structure and Nucleic acid tertiary structure, tertiary structure. Primary structur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
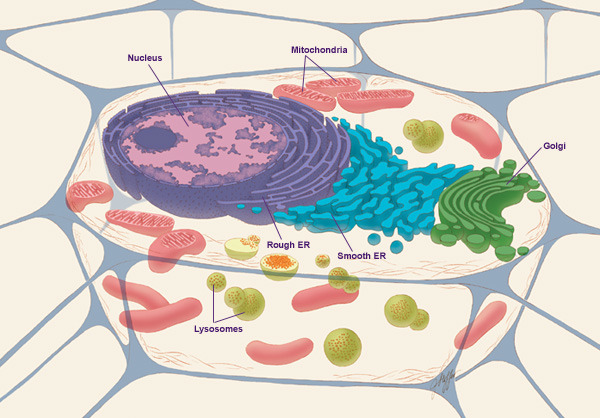
Cell Biology
Cell biology (also cellular biology or cytology) is a branch of biology that studies the structure, function, and behavior of cells. All living organisms are made of cells. A cell is the basic unit of life that is responsible for the living and functioning of organisms. Cell biology is the study of structural and functional units of cells. Cell biology encompasses both prokaryotic and eukaryotic cells and has many subtopics which may include the study of cell metabolism, cell communication, cell cycle, biochemistry, and cell composition. The study of cells is performed using several microscopy techniques, cell culture, and cell fractionation. These have allowed for and are currently being used for discoveries and research pertaining to how cells function, ultimately giving insight into understanding larger organisms. Knowing the components of cells and how cells work is fundamental to all biological sciences while also being essential for research in biomedical fields ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Dynamics
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics. Because molecular systems typically consist of a vast number of particles, it is impossible to determine the properties of such complex systems analytically; MD simulation circumvents this problem by using numerical methods. However, long MD simulations are mathematically ill-conditioned, generating cumulative err ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
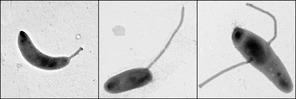
Caulobacter Crescentus
''Caulobacter crescentus'' is a Gram-negative, oligotrophic bacterium widely distributed in fresh water lakes and streams. The taxon is more properly known as ''Caulobacter vibrioides'' (Henrici and Johnson 1935). ''C. crescentus'' is an important model organism for studying the regulation of the cell cycle, asymmetric cell division, and cellular differentiation. ''Caulobacter'' daughter cells have two very different forms. One daughter is a mobile "swarmer" cell that has a single flagellum at one cell pole that provides swimming motility for chemotaxis. The other daughter, called the "stalked" cell, has a tubular stalk structure protruding from one pole that has an adhesive holdfast material on its end, with which the stalked cell can adhere to surfaces. Swarmer cells differentiate into stalked cells after a short period of motility. Chromosome replication and cell division only occurs in the stalked cell stage. ''C. crescentus'' derives its name from its crescent sh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |