|
Human Epigenome
Human epigenome is the complete set of structural modifications of chromatin and chemical modifications of histones and nucleotides (such as cytosine methylation). These modifications affect t according to cellular type and development status. Various studies show that epigenome depends on exogenous factors. Chemical modifications Different types of chemical modifications exist and the ChIP-seq experimental procedure can be performed in order to study them. The epigenetic profiles of human tissues reveals the following distinct histone modifications in different functional areas: Methylation DNA functionally interacts with a variety of epigenetic marks, such as cytosine methylation, also known as 5-methylcytosine (5mC). This epigenetic mark is widely conserved and plays major roles in the regulation of gene expression, in the silencing of transposable elements and repeat sequences. Individuals differ with their epigenetic profile, for example the variance in CpG methylatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenome
An epigenome consists of a record of the chemical changes to the DNA and histone proteins of an organism; these changes can be passed down to an organism's offspring via transgenerational stranded epigenetic inheritance. Changes to the epigenome can result in changes to the structure of chromatin and changes to the function of the genome. The epigenome is involved in regulating gene expression, development, tissue differentiation, and suppression of transposable elements. Unlike the underlying genome, which remains largely static within an individual, the epigenome can be dynamically altered by environmental conditions. Cancer Epigenetics is a currently active topic in cancer research. Human tumors undergo a major disruption of DNA methylation and histone modification patterns. The aberrant epigenetic landscape of the cancer cell is characterized by a global genomic hypomethylation, CpG island promoter hypermethylation of tumor suppressor genes, an altered histone code for c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
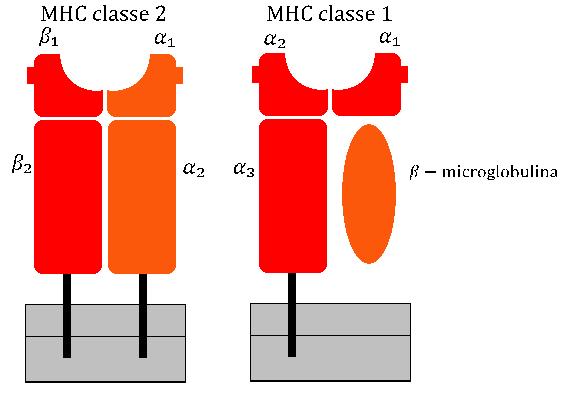
Major Histocompatibility Complex
The major histocompatibility complex (MHC) is a large locus on vertebrate DNA containing a set of closely linked polymorphic genes that code for cell surface proteins essential for the adaptive immune system. These cell surface proteins are called MHC molecules. This locus got its name because it was discovered via the study of transplanted tissue compatibility. Later studies revealed that tissue rejection due to incompatibility is only a facet of the full function of MHC molecules: binding an antigen derived from self-proteins, or from pathogens, and bringing the antigen presentation to the cell surface for recognition by the appropriate T-cells. MHC molecules mediate the interactions of leukocytes, also called white blood cells (WBCs), with other leukocytes or with body cells. The MHC determines donor compatibility for organ transplant, as well as one's susceptibility to autoimmune diseases. In a cell, protein molecules of the host's own phenotype or of other biologic entities ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fertilisation
Fertilisation or fertilization (see spelling differences), also known as generative fertilisation, syngamy and impregnation, is the fusion of gametes to give rise to a new individual organism or offspring and initiate its development. Processes such as insemination or pollination which happen before the fusion of gametes are also sometimes informally called fertilisation. The cycle of fertilisation and development of new individuals is called sexual reproduction. During double fertilisation in angiosperms the haploid male gamete combines with two haploid polar nuclei to form a triploid primary endosperm nucleus by the process of vegetative fertilisation. History In Antiquity, Aristotle conceived the formation of new individuals through fusion of male and female fluids, with form and function emerging gradually, in a mode called by him as epigenetic. In 1784, Spallanzani established the need of interaction between the female's ovum and male's sperm to form a zygote in frog ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Demethylation
Demethylation is the chemical process resulting in the removal of a methyl group (CH3) from a molecule. A common way of demethylation is the replacement of a methyl group by a hydrogen atom, resulting in a net loss of one carbon and two hydrogen atoms. The counterpart of demethylation is methylation. In biochemistry In biochemical systems, the process of demethylation is catalyzed by demethylases. These enzymes oxidize N-methyl groups, which occur in histones and some forms of DNA: :R2N-CH3 + O → R2N-H + CH2O One such oxidative enzyme family is the cytochrome P450. Alpha-ketoglutarate-dependent hydroxylases are active for demethylation of DNA, operating by a similar pathway. These reactions exploit the weak C-H bond adjacent to amines. In particular, 5-methylcytosines in DNA can be demethylated by TET enzymes as illustrated in the figure. TET enzymes are dioxygenases in the family of alpha-ketoglutarate-dependent hydroxylases. A TET enzyme is an alpha-ketoglut ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Immunostaining
In biochemistry, immunostaining is any use of an antibody-based method to detect a specific protein in a sample. The term "immunostaining" was originally used to refer to the immunohistochemical staining of tissue sections, as first described by Albert Coons in 1941. However, immunostaining now encompasses a broad range of techniques used in histology, cell biology, and molecular biology that use antibody-based staining methods. Techniques Immunohistochemistry Immunohistochemistry or IHC staining of tissue sections (or immunocytochemistry, which is the staining of cells), is perhaps the most commonly applied immunostaining technique. While the first cases of IHC staining used fluorescent dyes (see '' immunofluorescence''), other non-fluorescent methods using enzymes such as peroxidase (see '' immunoperoxidase staining'') and alkaline phosphatase are now used. These enzymes are capable of catalysing reactions that give a coloured product that is easily detectable by light m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulation Of Gene Expression
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from Transcriptional regulation, transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network. Gene regulation is essential for viruses, prokaryotes and eukaryotes as it increases the versatility and adaptability of an organism by allowing the cell to express protein when needed. Although as early as 1951, Barbara McClintock showed interaction between two genetic loci, Activator (''Ac'') and Dissociator (''Ds''), in the color f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcriptome
The transcriptome is the set of all RNA transcripts, including coding and non-coding, in an individual or a population of cells. The term can also sometimes be used to refer to all RNAs, or just mRNA, depending on the particular experiment. The term ''transcriptome'' is a portmanteau of the words ''transcript'' and ''genome''; it is associated with the process of transcript production during the biological process of transcription. The early stages of transcriptome annotations began with cDNA libraries published in the 1980s. Subsequently, the advent of high-throughput technology led to faster and more efficient ways of obtaining data about the transcriptome. Two biological techniques are used to study the transcriptome, namely DNA microarray, a hybridization-based technique and RNA-seq, a sequence-based approach. RNA-seq is the preferred method and has been the dominant transcriptomics technique since the 2010s. Single-cell transcriptomics allows tracking of transcript changes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Trans-acting
In the field of molecular biology, ''trans''-acting (''trans''-regulatory, ''trans''-regulation), in general, means "acting from a different molecule" (''i.e.'', intermolecular). It may be considered the opposite of ''cis''-acting (''cis''-regulatory, ''cis''-regulation), which, in general, means "acting from the same molecule" (''i.e.'', intramolecular). In the context of transcription regulation, a ''trans''-acting factor is usually a regulatory protein that binds to DNA. The binding of a ''trans''-acting factor to a ''cis''-regulatory element in DNA can cause changes in transcriptional expression levels. microRNAs or other diffusible molecules are also examples of ''trans''-acting factors that can regulate target sequences. The ''trans''-acting gene may be on a different chromosome to the target gene, but the activity is via the intermediary protein or RNA that it encodes. ''Cis''-acting elements, on the other hand, do not code for protein or RNA. Both the ''trans''-acti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymorphism (biology)
In biology, polymorphism is the occurrence of two or more clearly different morphs or forms, also referred to as alternative ''phenotypes'', in the population of a species. To be classified as such, morphs must occupy the same habitat at the same time and belong to a panmictic population (one with random mating). Ford E.B. 1965. ''Genetic polymorphism''. Faber & Faber, London. Put simply, polymorphism is when there are two or more possibilities of a trait on a gene. For example, there is more than one possible trait in terms of a jaguar's skin colouring; they can be light morph or dark morph. Due to having more than one possible variation for this gene, it is termed 'polymorphism'. However, if the jaguar has only one possible trait for that gene, it would be termed "monomorphic". For example, if there was only one possible skin colour that a jaguar could have, it would be termed monomorphic. The term polyphenism can be used to clarify that the different forms arise from the s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enhancer (genetics)
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are ''cis''-acting. They can be located up to 1 Mbp (1,000,000 bp) away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes. The first discovery of a eukaryotic enhancer was in the immunoglobulin heavy chain gene in 1983. This enhancer, located in the large intron, provided an explanation for the transcriptional activation of rearranged Vh gene promoters while unrearranged Vh promoters remained inactive. Locations In eukaryotic cells the structure of the chromatin complex of DNA is folded in a way that functionally mimics the supercoiled state characteristic of prokaryotic DNA, so although the en ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continue between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed; however, according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously turned over via RNA-induced transcriptional silencing (RITS). Recent studies with electron microscopy and OsO4 staining reveal that the dense packing is not due to the chromatin. Constitutive heterochromatin can affect the genes near itself (e.g. position-effect variegation). It is usually repetitive and forms structural functions such as centromeres or telomeres, in addition to acting as an attractor for other gene-expression or repression signals. Facultative hete ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |