|
Halomethanococcus
In taxonomy, the Methanosarcinaceae are a family of the Methanosarcinales. Phylogeny The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI) Biochemistry A notable trait of Methanosarcinaceae is that they are methanogens that incorporate the unusual amino acid pyrrolysine into their enzymes.Lehninger A, Nelson D, Cox M. Lehninger principles of biochemistry. 6th ed. New York: W.H. Freeman; 2013 p. 1124-1126. The enzyme monomethylamine methyltransferase catalyzes the reaction of monomethylamine to methane. This enzyme includes pyrrolysine. The unusual amino acid is inserted using a unique tRNA, the anticodon of which is UAG. In most organisms, and in most Methanosarcinaceae proteins, UAG is a stop codon. However in this enzyme, and anywhere else pyrrolysine is incorporated, likely through contextual markers on the mRNA, the pyrrolysine-loaded tRNA is inserted inste ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Halomethanococcus
In taxonomy, the Methanosarcinaceae are a family of the Methanosarcinales. Phylogeny The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI) Biochemistry A notable trait of Methanosarcinaceae is that they are methanogens that incorporate the unusual amino acid pyrrolysine into their enzymes.Lehninger A, Nelson D, Cox M. Lehninger principles of biochemistry. 6th ed. New York: W.H. Freeman; 2013 p. 1124-1126. The enzyme monomethylamine methyltransferase catalyzes the reaction of monomethylamine to methane. This enzyme includes pyrrolysine. The unusual amino acid is inserted using a unique tRNA, the anticodon of which is UAG. In most organisms, and in most Methanosarcinaceae proteins, UAG is a stop codon. However in this enzyme, and anywhere else pyrrolysine is incorporated, likely through contextual markers on the mRNA, the pyrrolysine-loaded tRNA is inserted inste ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Archaea Genera
This article lists the genera of the Archaea. The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI). Phylogeny National Center for Biotechnology Information (NCBI) taxonomy was initially used to decorate the genome tree via tax2tree. The 16S rRNA-based Greengenes taxonomy is used to supplement the taxonomy particularly in regions of the tree with no cultured representatives. List of Prokaryotic names with Standing in Nomenclature (LPSN) is used as the primary taxonomic authority for establishing naming priorities. Taxonomic ranks are normalised using phylorank and the taxonomy manually curated to remove polyphyletic groups. Cladogram was taken from the GTDB release 07-RS207 (8th April 2022). The position of clades with a "question mark" are based on the additional phylogeny of the 16S rRNA-based LTP_12_2021 by The All-Species Living Tree Project. Phylum " Altarcha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Family (biology)
Family ( la, familia, plural ') is one of the eight major hierarchical taxonomic ranks in Linnaean taxonomy. It is classified between order and genus. A family may be divided into subfamilies, which are intermediate ranks between the ranks of family and genus. The official family names are Latin in origin; however, popular names are often used: for example, walnut trees and hickory trees belong to the family Juglandaceae, but that family is commonly referred to as the "walnut family". What belongs to a family—or if a described family should be recognized at all—are proposed and determined by practicing taxonomists. There are no hard rules for describing or recognizing a family, but in plants, they can be characterized on the basis of both vegetative and reproductive features of plant species. Taxonomists often take different positions about descriptions, and there may be no broad consensus across the scientific community for some time. The publishing of new data and opini ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
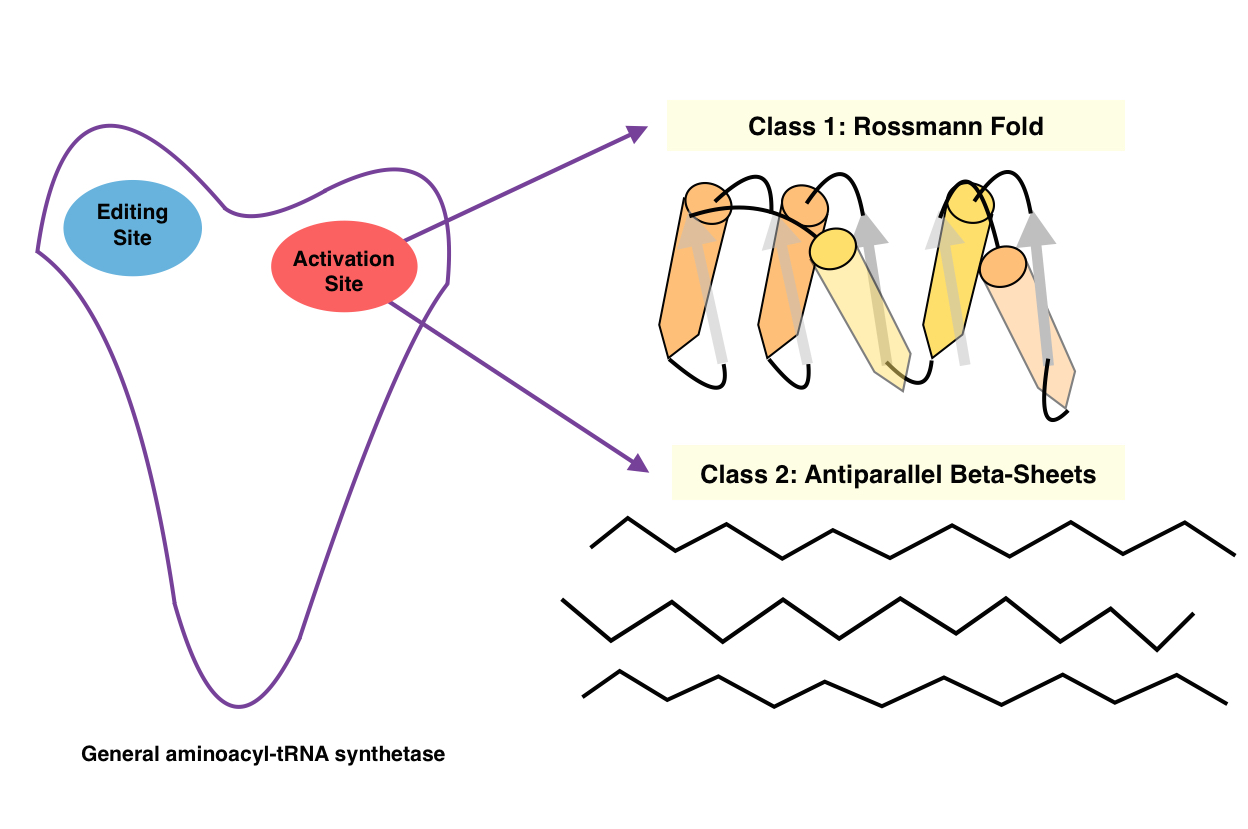
Aminoacyl-tRNA Synthetase
An aminoacyl-tRNA synthetase (aaRS or ARS), also called tRNA-ligase, is an enzyme that attaches the appropriate amino acid onto its corresponding tRNA. It does so by catalyzing the transesterification of a specific cognate amino acid or its precursor to one of all its compatible cognate tRNAs to form an aminoacyl-tRNA. In humans, the 20 different types of aa-tRNA are made by the 20 different aminoacyl-tRNA synthetases, one for each amino acid of the genetic code. This is sometimes called "charging" or "loading" the tRNA with an amino acid. Once the tRNA is charged, a ribosome can transfer the amino acid from the tRNA onto a growing peptide, according to the genetic code. Aminoacyl tRNA therefore plays an important role in RNA translation, the expression of genes to create proteins. Mechanism The synthetase first binds ATP and the corresponding amino acid (or its precursor) to form an aminoacyl-adenylate, releasing inorganic pyrophosphate (PPi). The adenylate-aaRS complex th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Release Factor
A release factor is a protein that allows for the termination of translation by recognizing the termination codon or stop codon in an mRNA sequence. They are named so because they release new peptides from the ribosome. Background During translation of mRNA, most codons are recognized by "charged" tRNA molecules, called aminoacyl-tRNAs because they are adhered to specific amino acids corresponding to each tRNA's anticodon. In the standard genetic code, there are three mRNA stop codons: UAG ("amber"), UAA ("ochre"), and UGA ("opal" or "umber"). Although these stop codons are triplets just like ordinary codons, they are not decoded by tRNAs. It was discovered by Mario Capecchi in 1967 that, instead, tRNAs do not ordinarily recognize stop codons at all, and that what he named "release factor" was not a tRNA molecule but a protein. Later, it was demonstrated that different release factors recognize different stop codons. Classification There are two classes of release factor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stop Codon
In molecular biology (specifically protein biosynthesis), a stop codon (or termination codon) is a codon (nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain. While start codons need nearby sequences or initiation factors to start translation, a stop codon alone is sufficient to initiate termination. Properties Standard codons In the standard genetic code, there are three different termination codons: Alternative stop codons There are variations on the standard genetic code, and alternative stop codons have been found in the mitochondrial genomes of vertebrates, ''Scenedesmus obliquus'', and '' Thra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TRNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino acid sequence of proteins. tRNAs genes from Bacteria are typically shorter (mean = 77.6 bp) than tRNAs from Archaea (mean = 83.1 bp) and eukaryotes (mean = 84.7 bp). The mature tRNA follows an opposite pattern with tRNAs from Bacteria being usually longer (median = 77.6 nt) than tRNAs from Archaea (median = 76.8 nt), with eukaryotes exhibiting the shortest mature tRNAs (median = 74.5 nt). Transfer RNA (tRNA) does this by carrying an amino acid to the protein synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methane
Methane ( , ) is a chemical compound with the chemical formula (one carbon atom bonded to four hydrogen atoms). It is a group-14 hydride, the simplest alkane, and the main constituent of natural gas. The relative abundance of methane on Earth makes it an economically attractive fuel, although capturing and storing it poses technical challenges due to its gaseous state under normal conditions for temperature and pressure. Naturally occurring methane is found both below ground and under the seafloor and is formed by both geological and biological processes. The largest reservoir of methane is under the seafloor in the form of methane clathrates. When methane reaches the surface and the atmosphere, it is known as atmospheric methane. The Earth's atmospheric methane concentration has increased by about 150% since 1750, and it accounts for 20% of the total radiative forcing from all of the long-lived and globally mixed greenhouse gases. It has also been detected on other plane ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monomethylamine
Methylamine is an organic compound with a formula of . This colorless gas is a derivative of ammonia, but with one hydrogen atom being replaced by a methyl group. It is the simplest primary amine. Methylamine is sold as a solution in methanol, ethanol, tetrahydrofuran, or water, or as the anhydrous gas in pressurized metal containers. Industrially, methylamine is transported in its anhydrous form in pressurized railcars and tank trailers. It has a strong odor similar to rotten fish. Methylamine is used as a building block for the synthesis of numerous other commercially available compounds. Industrial production Methylamine is prepared commercially by the reaction of ammonia with methanol in the presence of an aluminosilicate catalyst. Dimethylamine and trimethylamine are co-produced; the reaction kinetics and reactant ratios determine the ratio of the three products. The product most favored by the reaction kinetics is trimethylamine. : In this way, an estimated 115,000 tons ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monomethylamine Methyltransferase
Methylamine-corrinoid protein Co-methyltransferase (, ''mtmB (gene)'', ''monomethylamine methyltransferase'') is an enzyme with systematic name ''monomethylamine:5-hydroxybenzimidazolylcobamide Co-methyltransferase''. This enzyme catalyses the following chemical reaction : methylamine + o(I) methylamine-specific corrinoid protein\rightleftharpoons ethyl-Co(III) methylamine-specific corrinoid protein+ ammonia This enzyme is involved in methanogenesis Methanogenesis or biomethanation is the formation of methane coupled to energy conservation by microbes known as methanogens. Organisms capable of producing methane for energy conservation have been identified only from the domain Archaea, a group ... from methylamine. References External links * {{Portal bar, Biology, border=no EC 2.1.1 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyrrolysine
Pyrrolysine (symbol Pyl or O; encoded by the 'amber' stop codon UAG) is an α-amino acid that is used in the biosynthesis of proteins in some methanogenic archaea and bacteria; it is not present in humans. It contains an α-amino group (which is in the protonated – form under biological conditions), a carboxylic acid group (which is in the deprotonated –COO− form under biological conditions). Its pyrroline side-chain is similar to that of lysine in being basic and positively charged at neutral pH. Genetics Nearly all genes are translated using only 20 standard amino acid building blocks. Two unusual genetically-encoded amino acids are selenocysteine and pyrrolysine. Pyrrolysine was discovered in 2002 at the active site of methyltransferase enzyme from a methane-producing archeon, ''Methanosarcina barkeri''. This amino acid is encoded by UAG (normally a stop codon), and its synthesis and incorporation into protein is mediated via the biological machinery encoded by the ' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methanogens
Methanogens are microorganisms that produce methane as a metabolic byproduct in hypoxic conditions. They are prokaryotic and belong to the domain Archaea. All known methanogens are members of the archaeal phylum Euryarchaeota. Methanogens are common in wetlands, where they are responsible for marsh gas, and in the digestive tracts of animals such as ruminants and many humans, where they are responsible for the methane content of belching in ruminants and flatulence in humans. In marine sediments, the biological production of methane, also termed methanogenesis, is generally confined to where sulfates are depleted, below the top layers. Moreover, methanogenic archaea populations play an indispensable role in anaerobic wastewater treatments. Others are extremophiles, found in environments such as hot springs and submarine hydrothermal vents as well as in the "solid" rock of Earth's crust, kilometers below the surface. Physical description Methanogens are coccoid (spherical shaped) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |