|
HSUR
HSURs (''Herpesvirus saimiri'' U RNAs) are viral small regulatory RNAs. They are found in ''Herpesvirus saimiri'' which is responsible for aggressive T-cell leukemias in primates. They are nuclear RNAs which bind host proteins to form small nuclear ribonucleoproteins (snRNPs). The RNAs are 114–143 nucleotides in length and the HSUR family has been subdivided into HSURs numbered 1 to 7.Tycowski K.T., Kolev N.G., Conrad N.K., Fok V., and Steitz J.A. 2006. The ever-growing world of small nuclear ribonucleoproteins. In The RNA world, 3rd edition (ed. R.F. Gesteland et al.), p. 327. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. The function of HSURs has not yet been identified; they do not affect transcription so are thought to act post-transcriptionally, potentially influencing the stability of host mRNAs. HSUR1 and 2 are the most conserved members of the family within HSV subgroups. HSUR1 has been shown to bind the host heterogeneous ribonucleoprotein particl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Herpesvirus Saimiri
''Saimiriine gammaherpesvirus 2'' (SaHV-2) is a species of virus in the genus ''Rhadinovirus'', subfamily ''Gammaherpesvirinae'', family ''Herpesviridae'', and order ''Herpesvirales''. See also * HSUR (''Herpesvirus saimiri'' U RNAs) * Squirrel monkey Squirrel monkeys are New World monkeys of the genus ''Saimiri''. ''Saimiri'' is the only genus in the subfamily Saimirinae. The name of the genus is of Tupi origin (''sai-mirím'' or ''çai-mbirín'', with ''sai'' meaning 'monkey' and ''mirím'' ... (''Saimiri'') References External links * Gammaherpesvirinae {{Virus-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Herpesvirus Ateles
''Ateline gammaherpesvirus 2'' (AtHV-2) is a species of virus in the genus ''Rhadinovirus'', subfamily ''Gammaherpesvirinae ''Gammaherpesvirinae'' is a subfamily of viruses in the order ''Herpesvirales'' and in the family ''Herpesviridae''. Viruses in ''Gammaherpesvirinae'' are distinguished by reproducing at a more variable rate than other subfamilies of ''Herpesvir ...'', family '' Herpesviridae'', and order '' Herpesvirales''. See also * HSUR (''Herpesvirus saimiri'' U RNAs) * Spider monkey (''Ateles'') References External links * Gammaherpesvirinae {{Virus-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MiR-27
miR-27 is a family of microRNA precursors found in animals, including humans. MicroRNAs are typically transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. The excised region or, mature product, of the miR-27 precursor is the microRNA mir-27. ''Herpesvirus saimiri'' expresses several non-coding RNAs (HSURs) which have been found to significantly reduce the level of mir-27 in a host cell. It has been proposed that miR-27 operates together with miR-23 and mir-24 in a co-operative cluster. Regulation of adipocyte differentiation miR-27 is one of a number of microRNAs implicated in cholesterol homeostasis and fatty acid metabolism. The miR-27 gene family has been shown to be downregulated during the differentiation of adipocytes. miR-27 inhibits adipocyte formation when overexpressed, acting by blocking the expression of two main regulators of adipogenesis. MicroRNAs miR-27a and -27b have been found to negatively reg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase II
RNA polymerase II (RNAP II and Pol II) is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoters and begin transcription. Discovery Early studies suggested a minimum of two RNAPs: one which synthesized rRNA in the nucleolus, and one which synthesized other RNA in the nucleoplasm, part of the nucleus but outside the nucleolus. In 1969, science experimentalists Robert Roeder and William Rutter definitively discovered an additional RNAP that was responsible for transcription of some kind of RNA in the nucleoplasm. The finding was obtained by the use of ion-exchange chromatography via DEAE coated Sephadex beads. The technique separated the enzymes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HHV Latency Associated Transcript
HHV Latency Associated Transcript (HHV LAT) is a length of RNA which accumulates in cells hosting long-term, or ''latent'', Human Herpes Virus (HHV) infections. The LAT RNA is produced by genetic transcription from a certain region of the viral DNA. LAT regulates the viral genome and interferes with the normal activities of the infected host cell. Herpes virus may establish lifelong infection during which a ''reservoir'' virus population survives in host nerve cells for long periods of time. Such long-term Herpes infection requires a mode of cellular infection known as ''latent'' infection. During the latent infection, the metabolism of the host cell is disrupted. While the infected cell would ordinarily undergo an organized death or be removed by the immune system, the consequences of LAT production interfere with these normal processes. Latency is distinguished from ''lytic'' infection; in lytic infection many Herpes virus particles are produced and then burst or ''lyse' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MiRNA
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in mRNA molecules, then gene silence said mRNA molecules by one or more of the following processes: (1) cleavage of mRNA strand into two pieces, (2) destabilization of mRNA by shortening its poly(A) tail, or (3) translation of mRNA into proteins. This last method of gene silencing is the least efficient of the three, and requires the aid of ribosomes. miRNAs resemble the small interfering RNAs (siRNAs) of the RNA interference (RNAi) pathway, except miRNAs derive from regions of RNA transcripts that fold back on themselves to form short hairpins, whereas siRNAs derive from longer regions of double-stranded RNA. The human genome may encode over 1900 miRNAs, although more recent analysis suggests that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including ''co-suppression'', ''post-transcriptional gene silencing'' (PTGS), and ''quelling''. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm '' Caenorhabditis elegans'', which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sm Protein
In molecular biology, LSm proteins are a family of RNA-binding proteins found in virtually every cellular organism. LSm is a contraction of 'like Sm', because the first identified members of the LSm protein family were the Sm proteins. LSm proteins are defined by a characteristic three-dimensional structure and their assembly into rings of six or seven individual LSm protein molecules, and play a large number of various roles in mRNA processing and regulation. The Sm proteins were first discovered as antigens targeted by so-called anti-Sm antibodies in a patient with a form of systemic lupus erythematosus (SLE), a debilitating autoimmune disease. They were named Sm proteins in honor of Stephanie Smith, a patient who suffered from SLE. Other proteins with very similar structures were subsequently discovered and named LSm proteins. New members of the LSm protein family continue to be identified and reported. Proteins with similar structures are grouped into a hierarchy of p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many RNA secondary structures. As an important secondary structure of RNA, it can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA binding proteins, and serve as a substrate for enzymatic reactions. Formation and stability The formation of a stem-loop structure is dependent on the stability of the resulting helix and loop regions. The first prerequisite is the presence of a sequence that can fold back on itself to form a paired double helix. The stability of this helix is determined by its length, the number of mismatches or bulges it co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
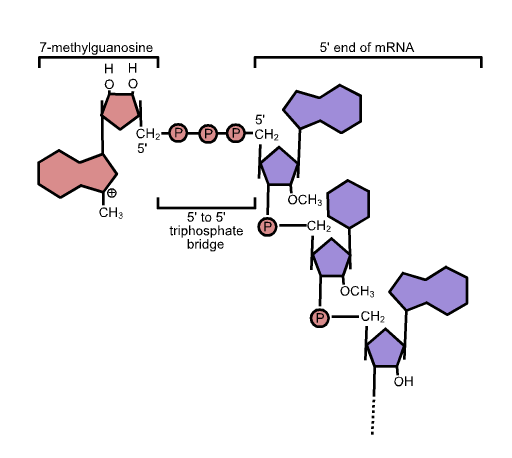
5′ Cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation of stable and mature messenger RNA able to undergo translation during protein synthesis. Mitochondrial mRNA and chloroplastic mRNA are not capped. Structure In eukaryotes, the 5′ cap (cap-0), found on the 5′ end of an mRNA molecule, consists of a guanine nucleotide connected to mRNA via an unusual 5′ to 5′ triphosphate linkage. This guanosine is methylated on the 7 position directly after capping ''in vivo'' by a methyltransferase. It is referred to as a 7-methylguanylate cap, abbreviated m7G. In multicellular eukaryotes and some viruses, further modifications exist, including the methylation of the 2′ hydroxy-groups of the first 2 ribose sugars of the 5′ end of the mRNA. cap-1 has a methylated 2′-hydroxy group ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (biology)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |