|
Genome Architecture Mapping
In molecular biology, genome architecture mapping (GAM) is a cryosectioning method to map colocalized DNA regions in a ligation independent manner. It overcomes some limitations of Chromosome conformation capture (3C), as these methods have a reliance on digestion and ligation to capture interacting DNA segments. GAM is the first genome-wide method for capturing three-dimensional proximities between any number of genomic loci without ligation. The sections that are found using the cryosectioning method mentioned above are referred to as “Nuclear Profiles”. The information that they provide relates to their coverage across a genome. A large set of values can be produced that represents the strength of nuclear profiles’ presence within a genome. Based on how large or small the coverage across a genome is, judgements can be made involving chromatin interactions, nuclear profile location within the nucleus being cryosectioned, and chromatin compaction levels. To be able to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology. Molecular biology was first described as an approach focused on the underpinnings of biological phenomena - uncovering the structures of biological molecules as well as their interactions, and how these interactions explain observations of classical biology. In 1945 the term molecular biology was used by physicist William Astbury. In 1953 Francis Crick, James Watson, Rosalind Franklin, and colleagues, working at Medical Research Council unit, Cavendish laboratory, Cambridge (now the MRC Laboratory of Molecular Biology), made a double helix model of DNA which changed the entire research scenario. They proposed the DNA structure based on previous research done by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cosegregation
Co-segregation is the likelihood of two units being inherited to the next generation or an interaction estimation probability between any number of loci. Interaction probability is determined using specified parts of a target gene (loci) and a group of nuclear profiles (NPs). The picture to the right serves to provide visual aid as to how a slice (NP) is taken from the nucleus and loci are searched for within the NP. Co-segregation used within other mathematical models (SLICE and Normalized Linkage Disequilibrium) assist in rendering 3-D visualizations as a smaller process of Genome Architecture Mapping (GAM). These renderings help determine genomic density and radial position. History Co-segregation in Genome Architecture Mapping (GAM) is a newer process being used to identify the compaction and adjacency of genomic windows. In a study from 2017, co-segregation was used to understand gene-expression-specific contacts in organizing the genome in mammalian nuclei in the lar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Next Generation Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with modern DN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
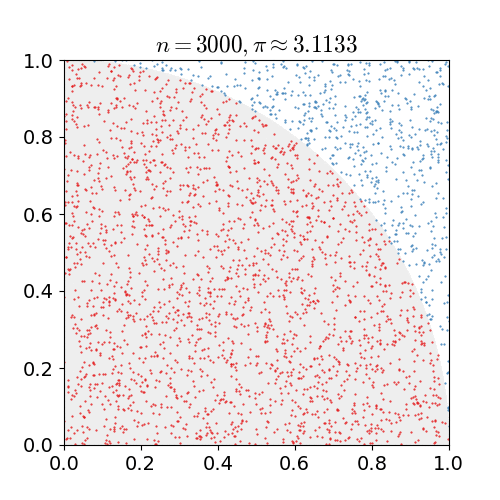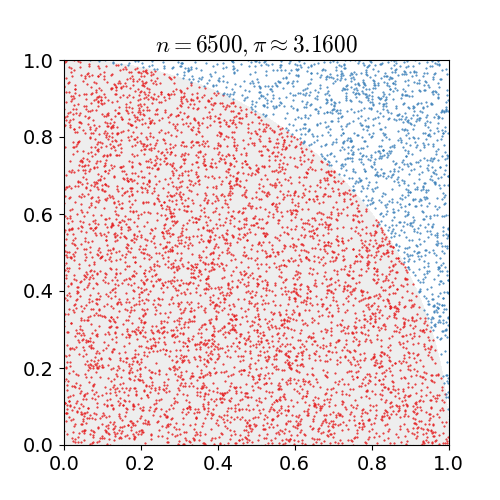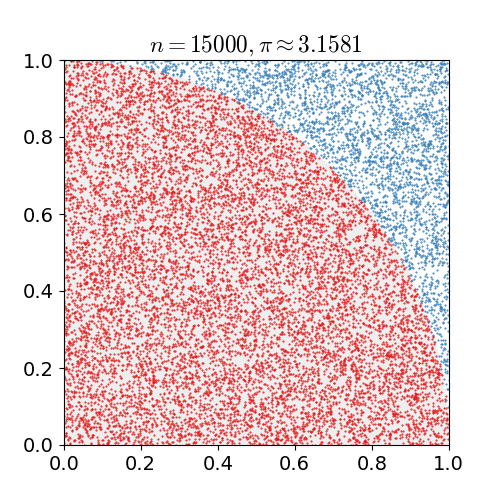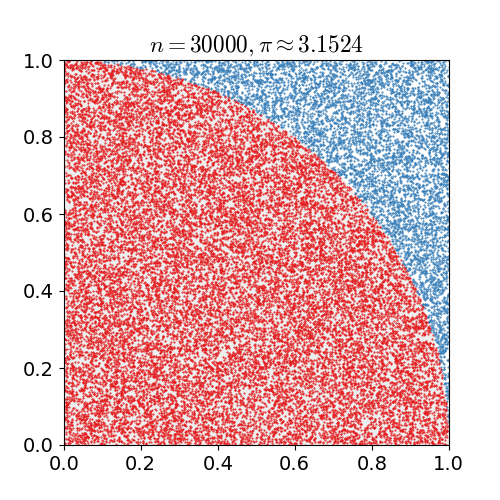
Monte Carlo Method
Monte Carlo methods, or Monte Carlo experiments, are a broad class of computational algorithms that rely on repeated random sampling to obtain numerical results. The underlying concept is to use randomness to solve problems that might be deterministic in principle. They are often used in physical and mathematical problems and are most useful when it is difficult or impossible to use other approaches. Monte Carlo methods are mainly used in three problem classes: optimization, numerical integration, and generating draws from a probability distribution. In physics-related problems, Monte Carlo methods are useful for simulating systems with many coupled degrees of freedom, such as fluids, disordered materials, strongly coupled solids, and cellular structures (see cellular Potts model, interacting particle systems, McKean–Vlasov processes, kinetic models of gases). Other examples include modeling phenomena with significant uncertainty in inputs such as the calculation of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Flowchart Of SLICE
A flowchart is a type of diagram that represents a workflow or process. A flowchart can also be defined as a diagrammatic representation of an algorithm, a step-by-step approach to solving a task. The flowchart shows the steps as boxes of various kinds, and their order by connecting the boxes with arrows. This diagrammatic representation illustrates a solution model to a given problem. Flowcharts are used in analyzing, designing, documenting or managing a process or program in various fields. * ''Document flowcharts'', showing controls over a document-flow through a system * ''Data flowcharts'', showing controls over a data-flow in a system * ''System flowcharts'', showing controls at a physical or resource level * ''Program flowchart'', showing the controls in a program within a system Notice that every type of flowchart focuses on some kind of control, rather than on the particular flow itself. However, there are some different classifications. For example, Andrew Veronis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eigenvector Centrality
In graph theory, eigenvector centrality (also called eigencentrality or prestige score) is a measure of the influence of a node in a network. Relative scores are assigned to all nodes in the network based on the concept that connections to high-scoring nodes contribute more to the score of the node in question than equal connections to low-scoring nodes. A high eigenvector score means that a node is connected to many nodes who themselves have high scores. Google's PageRank and the Katz centrality are variants of the eigenvector centrality. Using the adjacency matrix to find eigenvector centrality For a given graph G:=(V,E) with , V, vertices let A = (a_) be the adjacency matrix, i.e. a_ = 1 if vertex v is linked to vertex t, and a_ = 0 otherwise. The relative centrality score, x_v, of vertex v can be defined as: : x_v = \frac 1 \lambda \sum_ x_t = \frac 1 \lambda \sum_ a_ x_t where M(v) is the set of neighbors of v and \lambda is a constant. With a small rearrangement this ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Centrality
In graph theory and network analysis, indicators of centrality assign numbers or rankings to nodes within a graph corresponding to their network position. Applications include identifying the most influential person(s) in a social network, key infrastructure nodes in the Internet or urban networks, super-spreaders of disease, and brain networks. Centrality concepts were first developed in social network analysis, and many of the terms used to measure centrality reflect their sociological origin.Newman, M.E.J. 2010. ''Networks: An Introduction.'' Oxford, UK: Oxford University Press. Definition and characterization of centrality indices Centrality indices are answers to the question "What characterizes an important vertex?" The answer is given in terms of a real-valued function on the vertices of a graph, where the values produced are expected to provide a ranking which identifies the most important nodes. The word "importance" has a wide number of meanings, leading to man ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Clique Problem
In computer science, the clique problem is the computational problem of finding cliques (subsets of vertices, all adjacent to each other, also called complete subgraphs) in a graph. It has several different formulations depending on which cliques, and what information about the cliques, should be found. Common formulations of the clique problem include finding a maximum clique (a clique with the largest possible number of vertices), finding a maximum weight clique in a weighted graph, listing all maximal cliques (cliques that cannot be enlarged), and solving the decision problem of testing whether a graph contains a clique larger than a given size. The clique problem arises in the following real-world setting. Consider a social network, where the graph's vertices represent people, and the graph's edges represent mutual acquaintance. Then a clique represents a subset of people who all know each other, and algorithms for finding cliques can be used to discover these groups of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enhancer (genetics)
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are ''cis''-acting. They can be located up to 1 Mbp (1,000,000 bp) away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes. The first discovery of a eukaryotic enhancer was in the immunoglobulin heavy chain gene in 1983. This enhancer, located in the large intron, provided an explanation for the transcriptional activation of rearranged Vh gene promoters while unrearranged Vh promoters remained inactive. Locations In eukaryotic cells the structure of the chromatin complex of DNA is folded in a way that functionally mimics the supercoiled state characteristic of prokaryotic DNA, so although the en ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NP-hardness
In computational complexity theory, NP-hardness ( non-deterministic polynomial-time hardness) is the defining property of a class of problems that are informally "at least as hard as the hardest problems in NP". A simple example of an NP-hard problem is the subset sum problem. A more precise specification is: a problem ''H'' is NP-hard when every problem ''L'' in NP can be reduced in polynomial time to ''H''; that is, assuming a solution for ''H'' takes 1 unit time, ''H''s solution can be used to solve ''L'' in polynomial time. As a consequence, finding a polynomial time algorithm to solve any NP-hard problem would give polynomial time algorithms for all the problems in NP. As it is suspected that P≠NP, it is unlikely that such an algorithm exists. It is suspected that there are no polynomial-time algorithms for NP-hard problems, but that has not been proven. Moreover, the class P, in which all problems can be solved in polynomial time, is contained in the NP class. Defi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |