|
Gene Silencing
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene. Gene silencing can occur during either transcription or translation and is often used in research. In particular, methods used to silence genes are being increasingly used to produce therapeutics to combat cancer and other diseases, such as infectious diseases and neurodegenerative disorders. Gene silencing is often considered the same as gene knockdown. When genes are silenced, their expression is reduced. In contrast, when genes are knocked out, they are completely erased from the organism's genome and, thus, have no expression. Gene silencing is considered a gene knockdown mechanism since the methods used to silence genes, such as RNAi, CRISPR, or siRNA, generally reduce the expression of a gene by at least 70% but do not eliminate it. Methods using gene silencing are often considered better than gene knockouts since they allow researchers to study essential genes that are r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulation Of Gene Expression
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from Transcriptional regulation, transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network. Gene regulation is essential for viruses, prokaryotes and eukaryotes as it increases the versatility and adaptability of an organism by allowing the cell to express protein when needed. Although as early as 1951, Barbara McClintock showed interaction between two genetic loci, Activator (''Ac'') and Dissociator (''Ds''), in the color f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-directed DNA Methylation
RNA-directed DNA methylation (RdDM) is a biological process in which non-coding RNA molecules direct the addition of DNA methylation to specific DNA sequences. The RdDM pathway is unique to plants, although other mechanisms of RNA-directed chromatin modification have also been described in fungi and animals. To date, the RdDM pathway is best characterized within angiosperms (flowering plants), and particularly within the model plant ''Arabidopsis thaliana''. However, conserved RdDM pathway components and associated small RNAs (sRNAs) have also been found in other groups of plants, such as gymnosperms and ferns. The RdDM pathway closely resembles other sRNA pathways, particularly the highly conserved RNAi pathway found in fungi, plants, and animals. Both the RdDM and RNAi pathways produce sRNAs and involve conserved Argonaute, Dicer and RNA-dependent RNA polymerase proteins. RdDM has been implicated in a number of regulatory processes in plants. The DNA methylation added by RdDM ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
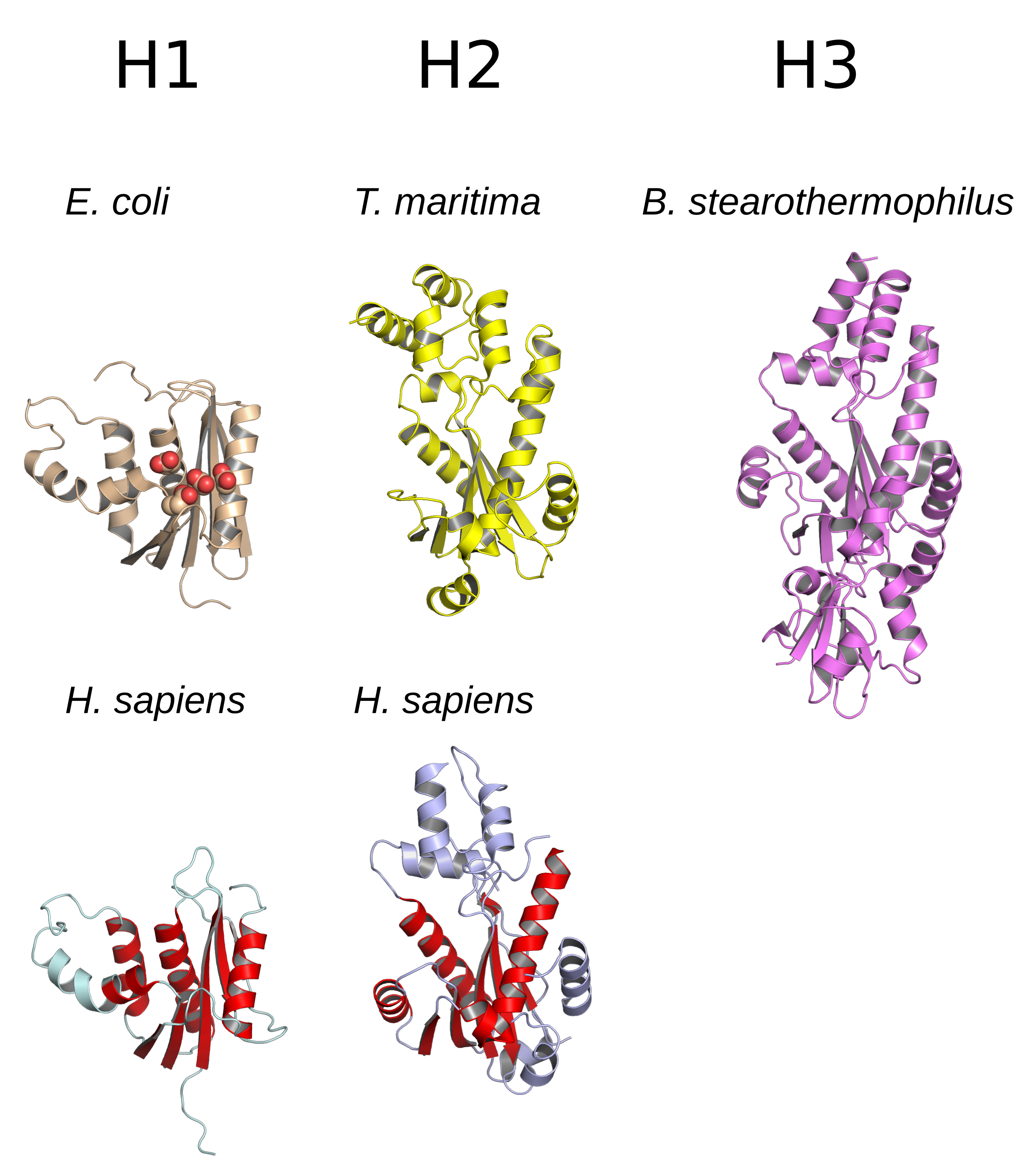
Ribozyme Mechanism
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or in vitro-evolved ribozymes are the cleavage or ligation of RNA and DNA and peptide bond formation. For example, the smallest ribozyme known (GUGGC-3') can aminoacylate a GCCU-3' sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during protein synthesis. They also participate in a variety of RNA processing reactions, including RNA splicing, viral replicati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heteroduplex
A heteroduplex is a double-stranded ( duplex) molecule of nucleic acid originated through the genetic recombination of single complementary strands derived from ''different'' sources, such as from different homologous chromosomes or even from different organisms. One such example is the heteroduplex DNA strand formed in hybridization processes, usually for biochemistry-based phylogenetic analyses. Another is the heteroduplexes formed when non-natural analogs of nucleic acids are used to bind with nucleic acids; these heteroduplexes result from performing antisense techniques using single-stranded peptide nucleic acid, 2'-O-methyl phosphorothioate or Morpholino oligos to bind with RNA. In meiosis, the process of crossing-over occurs between non-sister chromatids, which results in new allelic combinations in the gametes. In crossing-over, a Spo11 enzyme makes staggered nicks in a pair of sister chromatid strands (in a tetrad organization of prophase). Subsequent enzymes tri ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligonucleotides
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression (e.g. microRNA), or are degradation intermediates derived from the breakdown of larger nucleic acid molecules. Oligonucleotides are characterized by the sequence of nucleotide residues that make up the entire molecule. The length of the oligonucleotide is usually denoted by " -mer" (from Greek ''meros'', "part"). For example, an oligonucleotide of six nucleotides (nt) is a hexamer, while one of 25 nt wou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein. mRNA is created during the process of Transcription (biology), transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as Translation (biology), translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genet ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase H
Ribonuclease H (abbreviated RNase H or RNH) is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/ DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes. The family is divided into evolutionarily related groups with slightly different substrate preferences, broadly designated ribonuclease H1 and H2. The human genome encodes both H1 and H2. Human ribonuclease H2 is a heterotrimeric complex composed of three subunits, mutations in any of which are among the genetic causes of a rare disease known as Aicardi–Goutières syndrome. A third type, closely related to H2, is found only in a few prokaryotes, whereas H1 and H2 occur in all domains of life. Additionally, RNase H1-like retroviral ribonuclease H domains occur in multidomain reverse transcriptase proteins, which are encoded by retroviruses such as HIV and are required for viral repl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). If the sugar is ribose, the polymer is RNA; if the sugar is the ribose derivative deoxyribose, the polymer is DNA. Nucleic acids are naturally occurring chemical compounds that serve as the primary information-carrying molecules in cells and make up the genetic material. Nucleic acids are found in abundance in all living things, where they create, encode, and then store information of every living cell of every life-form on Earth. In turn, they function to transmit and express that information inside and outside the cell nucleus to the interior operations of the cell and ultimately to the next generation of each living organism. The encoded information is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligonucleotides
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression (e.g. microRNA), or are degradation intermediates derived from the breakdown of larger nucleic acid molecules. Oligonucleotides are characterized by the sequence of nucleotide residues that make up the entire molecule. The length of the oligonucleotide is usually denoted by " -mer" (from Greek ''meros'', "part"). For example, an oligonucleotide of six nucleotides (nt) is a hexamer, while one of 25 nt wou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Paul Zamecnik
Paul Charles Zamecnik (November 22, 1912 – October 27, 2009) was an American scientist who played a central role in the early history of molecular biology. He was a professor of medicine at Harvard Medical School and a senior scientist at Massachusetts General Hospital. Zamecnik pioneered the in vitro synthesis of proteins and helped elucidate the way cells generate proteins. With Mahlon Hoagland he co-discovered transfer RNA (tRNA). Through his later work, he is credited as the inventor of antisense therapeutics. Throughout his career, Zamecnik earned over a dozen US patents for his therapeutic techniques. Up until his death in 2009 he maintained a lab at MGH where he studied the application of synthetic oligonucleotides (antisense hybrids) for chemotherapeutic treatment of drug resistant and XDR tuberculosis in his later years. Education and research Paul Zamecnik was born in Cleveland, Ohio to John Charles Zamecnik (1879-1930) and Mary Gertrude Mccarthy (1883-1937). John ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antisense Oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression (e.g. microRNA), or are degradation intermediates derived from the breakdown of larger nucleic acid molecules. Oligonucleotides are characterized by the sequence of nucleotide residues that make up the entire molecule. The length of the oligonucleotide is usually denoted by "-mer" (from Greek ''meros'', "part"). For example, an oligonucleotide of six nucleotides (nt) is a hexamer, while one of 25 nt woul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transvection (genetics)
Transvection is an epigenetic phenomenon that results from an interaction between an allele on one chromosome and the corresponding allele on the homologous chromosome. Transvection can lead to either gene activation or repression. It can also occur between nonallelic regions of the genome as well as regions of the genome that are not transcribed. The first observation of mitotic (i.e. non-meiotic) chromosome pairing was discovered via microscopy in 1908 by Nettie Stevens. Edward B. Lewis at Caltech discovered transvection at the bithorax complex in Drosophila in the 1950s. Since then, transvection has been observed at a number of additional loci in Drosophila, including white, decapentaplegic, eyes absent, vestigial, and yellow. As stated by Ed Lewis, "Operationally, transvection is occurring if the phenotype of a given genotype can be altered solely by disruption of somatic (or meiotic) pairing. Such disruption can generally be accomplished by introduction of a heterozygous rear ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |