|
Gene Knockouts
A gene knockout (abbreviation: KO) is a genetic technique in which one of an organism's genes is made inoperative ("knocked out" of the organism). However, KO can also refer to the gene that is knocked out or the organism that carries the gene knockout. Knockout organisms or simply knockouts are used to study gene function, usually by investigating the effect of gene loss. Researchers draw inferences from the difference between the knockout organism and normal individuals. The KO technique is essentially the opposite of a gene knock-in. Knocking out two genes simultaneously in an organism is known as a double knockout (DKO). Similarly the terms triple knockout (TKO) and quadruple knockouts (QKO) are used to describe three or four knocked out genes, respectively. However, one needs to distinguish between heterozygous and homozygous KOs. In the former, only one of two gene copies (alleles) is knocked out, in the latter both are knocked out. Methods Knockouts are accomplished throu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetics
Genetics is the study of genes, genetic variation, and heredity in organisms.Hartl D, Jones E (2005) It is an important branch in biology because heredity is vital to organisms' evolution. Gregor Mendel, a Moravian Augustinian friar working in the 19th century in Brno, was the first to study genetics scientifically. Mendel studied "trait inheritance", patterns in the way traits are handed down from parents to offspring over time. He observed that organisms (pea plants) inherit traits by way of discrete "units of inheritance". This term, still used today, is a somewhat ambiguous definition of what is referred to as a gene. Trait inheritance and molecular inheritance mechanisms of genes are still primary principles of genetics in the 21st century, but modern genetics has expanded to study the function and behavior of genes. Gene structure and function, variation, and distribution are studied within the context of the cell, the organism (e.g. dominance), and within the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Physcomitrella Knockout Mutants
''Physcomitrella'' is a genus of mosses, containing two species. ''Physcomitrella patens'' is a model organism in laboratory research. '' Physcomitrella readeri'' is fairly similar, distinguished only by subtle characteristics. In fact, it has often been debated whether they should rightly be considered separate species, or a single species. ''Physcomitrella'' has been used as a model organism. It diverged from the flowering plants over 400 million years ago, so the difference between the groups indicates the changes in the genome In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding g ... that accompanied the adaptation of plants to living on land. References Moss genera Funariales {{Bryophyte-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Guide RNA
A guide RNA (gRNA) is a piece of RNA that functions as a guide for RNA- or DNA-targeting enzymes, with which it forms complexes. Very often these enzymes will delete, insert or otherwise alter the targeted RNA or DNA. They occur naturally, serving important functions, but can also be designed to be used for targeted editing, such as with CRISPR-Cas9 and CRISPR-Cas12. History RNA-editing Guide RNA was discovered in 1990 by B. Blum, N. Bakalara, and L. Simpson in the mitochondria of protists called Leishmania tarentolae. The guide RNA there is encoded in maxicircle DNA and contains sequences matching those within the edited regions of the mRNA. They enable the cleavage, insertion and deletion of bases. Guide RNA in Protists Trypanosomatid protists and other kinetoplastids have a novel post-transcriptional mitochondrial RNA modification process known as "RNA editing". They have a large segment of highly organized DNA segments in their mitochondria. This mitochondrial DNA is ci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CRISPR
CRISPR () (an acronym for clustered regularly interspaced short palindromic repeats) is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote. They are used to detect and destroy DNA from similar bacteriophages during subsequent infections. Hence these sequences play a key role in the antiviral (i.e. anti-phage) defense system of prokaryotes and provide a form of acquired immunity. CRISPR is found in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea. Cas9 (or "CRISPR-associated protein 9") is an enzyme that uses CRISPR sequences as a guide to recognize and cleave specific strands of DNA that are complementary to the CRISPR sequence. Cas9 enzymes together with CRISPR sequences form the basis of a technology known as CRISPR-Cas9 that can be used to edit genes within organisms. This editi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TALENs
Transcription activator-like effector nucleases (TALEN) are restriction enzymes that can be engineered to cut specific sequences of DNA. They are made by fusing a TAL effector DNA-binding domain to a DNA cleavage domain (a nuclease which cuts DNA strands). Transcription activator-like effectors (TALEs) can be engineered to bind to practically any desired DNA sequence, so when combined with a nuclease, DNA can be cut at specific locations. The restriction enzymes can be introduced into cells, for use in gene editing or for genome editing ''in situ'', a technique known as genome editing with engineered nucleases. Alongside zinc finger nucleases and CRISPR/Cas9, TALEN is a prominent tool in the field of genome editing. TALE DNA-binding domain TAL effectors are proteins that are secreted by ''Xanthomonas'' bacteria via their type III secretion system when they infect plants. The DNA binding domain contains a repeated highly conserved 33–34 amino acid sequence with divergent 12t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone (i.e. each strand) of the DNA double helix. These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses. Inside a prokaryote, the restriction enzymes selectively cut up ''foreign'' DNA in a process called ''restriction digestion''; meanwhile, host DNA is protected by a modification enzyme (a methyltransferase) that mod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Zinc Finger Nuclease
Zinc-finger nucleases (ZFNs) are artificial restriction enzymes generated by fusing a zinc finger DNA-binding domain to a DNA-cleavage domain. Zinc finger domains can be engineered to target specific desired DNA sequences and this enables zinc-finger nucleases to target unique sequences within complex genomes. By taking advantage of endogenous DNA repair machinery, these reagents can be used to precisely alter the genomes of higher organisms. Alongside CRISPR/Cas9 and TALEN, ZFN is a prominent tool in the field of genome editing. Domains DNA-binding domain The DNA-binding domains of individual ZFNs typically contain between three and six individual zinc finger repeats and can each recognize between 9 and 18 basepairs. If the zinc finger domains perfectly recognize a 3 basepair DNA sequence, they can generate a 3-finger array that can recognize a 9 basepair target site. Other procedures can utilize either 1-finger or 2-finger modules to generate zinc-finger arrays with s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
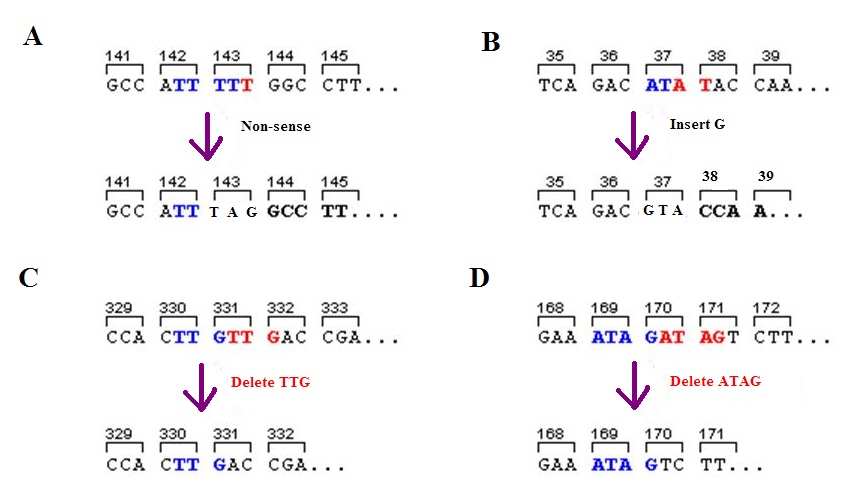
Frameshift Mutation
A frameshift mutation (also called a framing error or a reading frame shift) is a genetic mutation caused by indels ( insertions or deletions) of a number of nucleotides in a DNA sequence that is not divisible by three. Due to the triplet nature of gene expression by codons, the insertion or deletion can change the reading frame (the grouping of the codons), resulting in a completely different translation from the original. The earlier in the sequence the deletion or insertion occurs, the more altered the protein. A frameshift mutation is not the same as a single-nucleotide polymorphism in which a nucleotide is replaced, rather than inserted or deleted. A frameshift mutation will in general cause the reading of the codons after the mutation to code for different amino acids. The frameshift mutation will also alter the first stop codon ("UAA", "UGA" or "UAG") encountered in the sequence. The polypeptide being created could be abnormally short or abnormally long, and will most l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-homologous End Joining
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair(HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber. NHEJ is typically guided by short homologous DNA sequences called microhomologies. These microhomologies are often present in single-stranded overhangs on the ends of double-strand breaks. When the overhangs are perfectly compatible, NHEJ usually repairs the break accurately. Imprecise repair leading to loss of nucleotides can also occur, but is much more common when the overhangs are not compatible. Inappropriate NHEJ can lead to translocations and telomere fusion, hallma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Frameshift Mutations (13080927393)
A frameshift mutation (also called a framing error or a reading frame shift) is a genetic mutation caused by indels ( insertions or deletions) of a number of nucleotides in a DNA sequence that is not divisible by three. Due to the triplet nature of gene expression by codons, the insertion or deletion can change the reading frame (the grouping of the codons), resulting in a completely different translation from the original. The earlier in the sequence the deletion or insertion occurs, the more altered the protein. A frameshift mutation is not the same as a single-nucleotide polymorphism in which a nucleotide is replaced, rather than inserted or deleted. A frameshift mutation will in general cause the reading of the codons after the mutation to code for different amino acids. The frameshift mutation will also alter the first stop codon ("UAA", "UGA" or "UAG") encountered in the sequence. The polypeptide being created could be abnormally short or abnormally long, and will most li ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homozygous
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism. Most eukaryotes have two matching sets of chromosomes; that is, they are diploid. Diploid organisms have the same loci on each of their two sets of homologous chromosomes except that the sequences at these loci may differ between the two chromosomes in a matching pair and that a few chromosomes may be mismatched as part of a chromosomal sex-determination system. If both alleles of a diploid organism are the same, the organism is homozygous at that locus. If they are different, the organism is heterozygous at that locus. If one allele is missing, it is hemizygous, and, if both alleles are missing, it is nullizygous. The DNA sequence of a gene often varies from one individual to another. These gene variants are called alleles. While so ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Selective Breeding
Selective breeding (also called artificial selection) is the process by which humans use animal breeding and plant breeding to selectively develop particular phenotypic traits (characteristics) by choosing which typically animal or plant males and females will sexually reproduce and have offspring together. Domesticated animals are known as breeds, normally bred by a professional breeder, while domesticated plants are known as varieties, cultigens, cultivars, or breeds. Two purebred animals of different breeds produce a crossbreed, and crossbred plants are called hybrids. Flowers, vegetables and fruit-trees may be bred by amateurs and commercial or non-commercial professionals: major crops are usually the provenance of the professionals. In animal breeding, techniques such as inbreeding, linebreeding, and outcrossing are utilized. In plant breeding, similar methods are used. Charles Darwin discussed how selective breeding had been successful in producing change o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |