|
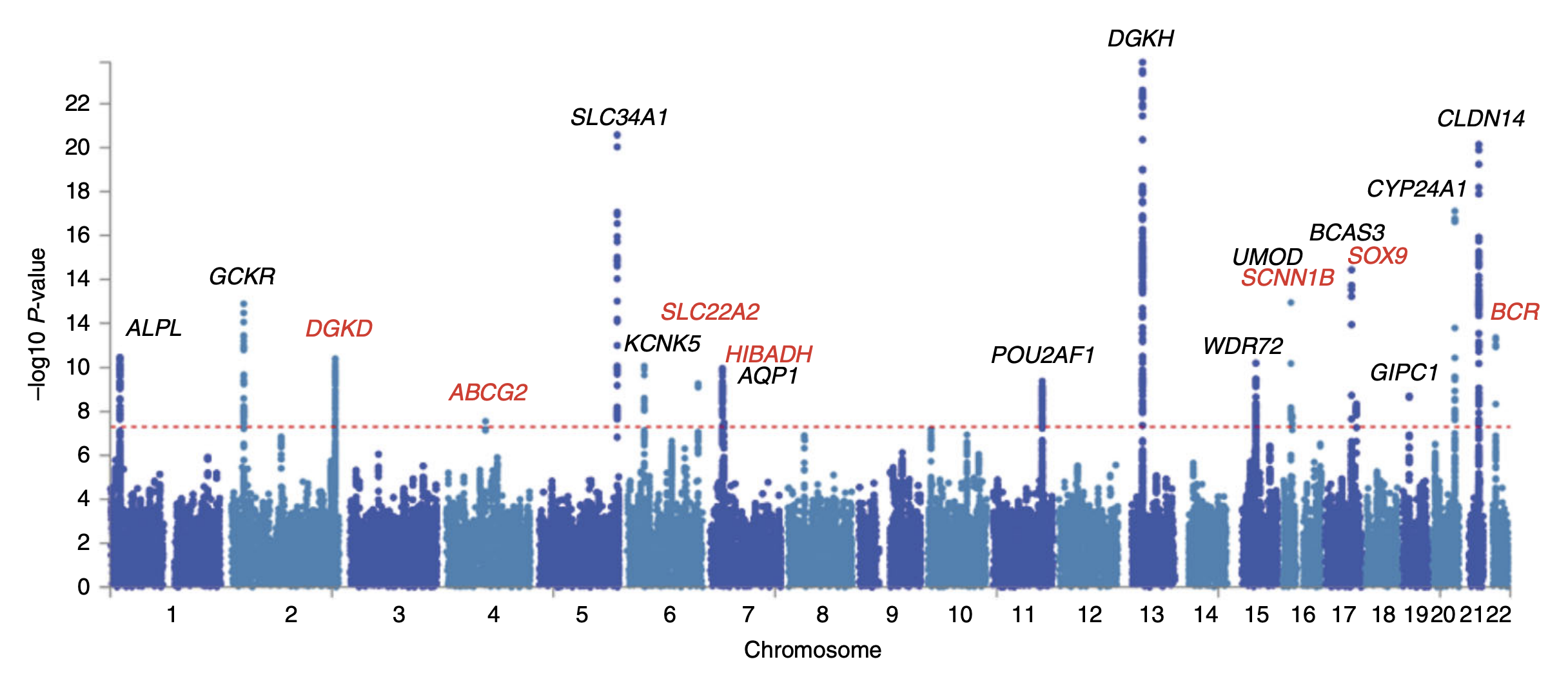
GWAS
In genomics, a genome-wide association study (GWA study, or GWAS), also known as whole genome association study (WGA study, or WGAS), is an observational study of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms. When applied to human data, GWA studies compare the DNA of participants having varying phenotypes for a particular trait or disease. These participants may be people with a disease (cases) and similar people without the disease (controls), or they may be people with different phenotypes for a particular trait, for example blood pressure. This approach is known as phenotype-first, in which the participants are classified first by their clinical manifestation(s), as opposed to genotype-first. Each pers ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genomics
Genomics is an interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, three-dimensional structural configuration. In contrast to genetics, which refers to the study of ''individual'' genes and their roles in inheritance, genomics aims at the collective characterization and quantification of ''all'' of an organism's genes, their interrelations and influence on the organism. Genes may direct the production of proteins with the assistance of enzymes and messenger molecules. In turn, proteins make up body structures such as organs and tissues as well as control chemical reactions and carry signals between cells. Genomics also involves the sequencing and analysis of genomes through uses of high throughput DNA sequencing and bioinformatics to assemble and analyze the function and structure of entire genomes. Advances in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
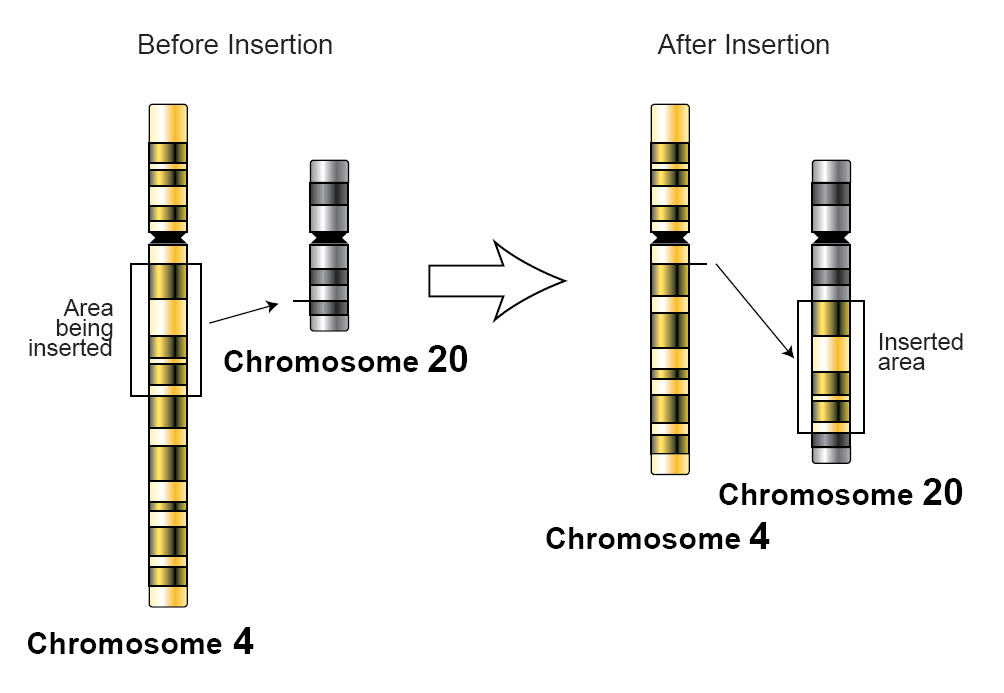
Insertion (genetics)
In genetics, an insertion (also called an insertion mutation) is the addition of one or more nucleotide base pairs into a DNA sequence. This can often happen in microsatellite regions due to the DNA polymerase slipping. Insertions can be anywhere in size from one base pair incorrectly inserted into a DNA sequence to a section of one chromosome inserted into another. The mechanism of the smallest single base insertion mutations is believed to be through base-pair separation between the template and primer strands followed by non-neighbor base stacking, which can occur locally within the DNA polymerase active site. On a chromosome level, an ''insertion'' refers to the insertion of a larger sequence into a chromosome. This can happen due to unequal crossover during meiosis. N region addition is the addition of non-coded nucleotides during recombination by terminal deoxynucleotidyl transferase. P nucleotide insertion is the insertion of palindromic sequences encoded by the ends ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chi-squared Test
A chi-squared test (also chi-square or test) is a statistical hypothesis test used in the analysis of contingency tables In statistics, a contingency table (also known as a cross tabulation or crosstab) is a type of table in a matrix format that displays the (multivariate) frequency distribution of the variables. They are heavily used in survey research, business ... when the sample sizes are large. In simpler terms, this test is primarily used to examine whether two categorical variables (''two dimensions of the contingency table'') are independent in influencing the test statistic (''values within the table''). The test is Validity (statistics), valid when the test statistic is chi-squared distribution, chi-squared distributed under the null hypothesis, specifically Pearson's chi-squared test and variants thereof. Pearson's chi-squared test is used to determine whether there is a Statistical significance, statistically significant difference between the expected frequency ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
P-value
In null-hypothesis significance testing, the ''p''-value is the probability of obtaining test results at least as extreme as the result actually observed, under the assumption that the null hypothesis is correct. A very small ''p''-value means that such an extreme observed outcome would be very unlikely under the null hypothesis. Reporting ''p''-values of statistical tests is common practice in academic publications of many quantitative fields. Since the precise meaning of ''p''-value is hard to grasp, misuse is widespread and has been a major topic in metascience. Basic concepts In statistics, every conjecture concerning the unknown probability distribution of a collection of random variables representing the observed data X in some study is called a ''statistical hypothesis''. If we state one hypothesis only and the aim of the statistical test is to see whether this hypothesis is tenable, but not to investigate other specific hypotheses, then such a test is called a null ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Odds Ratio
An odds ratio (OR) is a statistic that quantifies the strength of the association between two events, A and B. The odds ratio is defined as the ratio of the odds of A in the presence of B and the odds of A in the absence of B, or equivalently (due to symmetry), the ratio of the odds of B in the presence of A and the odds of B in the absence of A. Two events are independent if and only if the OR equals 1, i.e., the odds of one event are the same in either the presence or absence of the other event. If the OR is greater than 1, then A and B are associated (correlated) in the sense that, compared to the absence of B, the presence of B raises the odds of A, and symmetrically the presence of A raises the odds of B. Conversely, if the OR is less than 1, then A and B are negatively correlated, and the presence of one event reduces the odds of the other event. Note that the odds ratio is symmetric in the two events, and there is no causal direction implied ( correlation does not imply c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
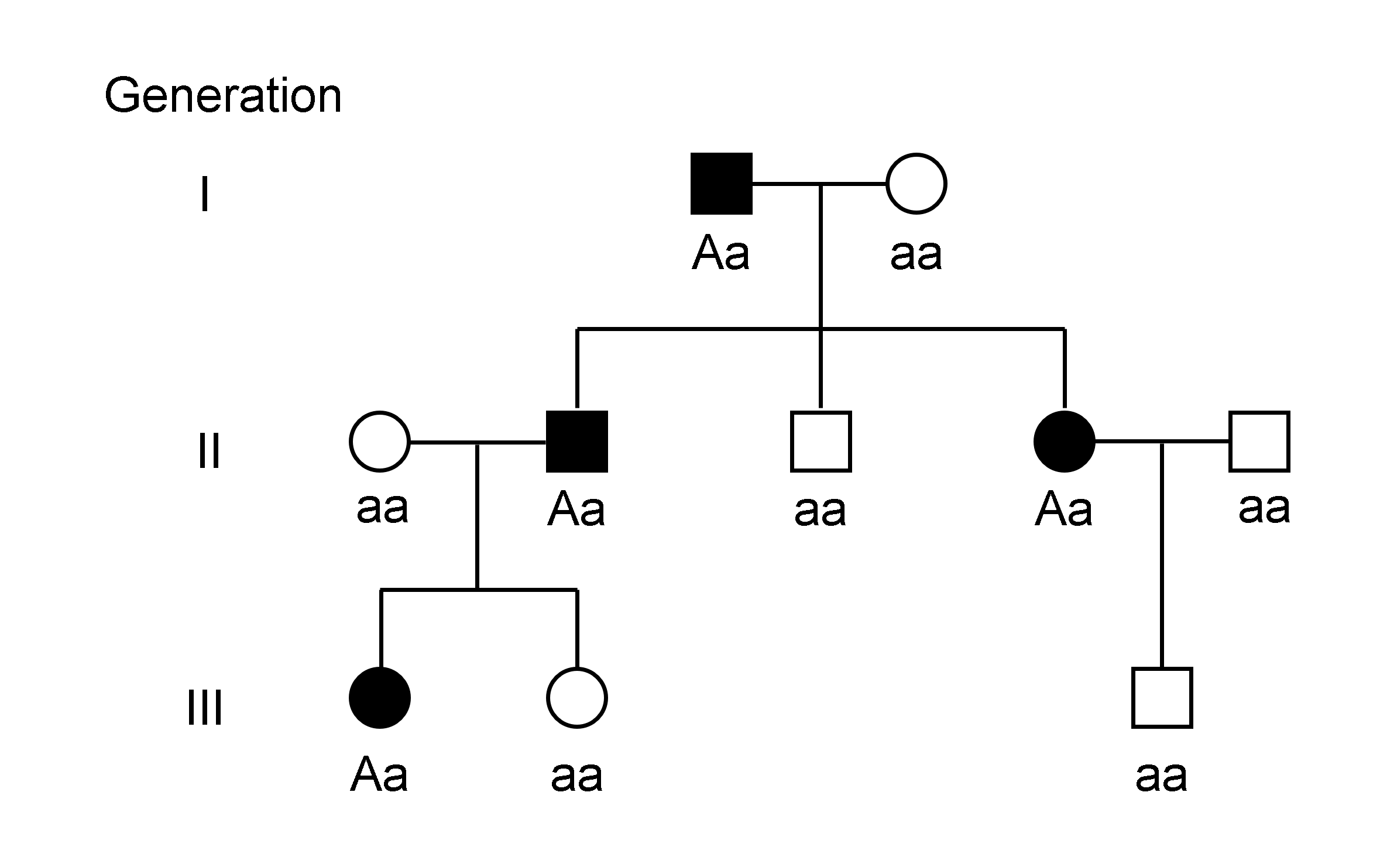
Genotype Phenotype Regression
The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a specific gene depends on the number of copies of each chromosome found in that species, also referred to as ploidy. In diploid species like humans, two full sets of chromosomes are present, meaning each individual has two alleles for any given gene. If both alleles are the same, the genotype is referred to as homozygous. If the alleles are different, the genotype is referred to as heterozygous. Genotype contributes to phenotype, the observable traits and characteristics in an individual or organism. The degree to which genotype affects phenotype depends on the trait. For example, the petal color in a pea plant is exclusively determined by genotype. The petals can be purple or white depending on the alleles present in the pea plant. Howeve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Method Example For GWA Study Designs
Method ( grc, μέθοδος, methodos) literally means a pursuit of knowledge, investigation, mode of prosecuting such inquiry, or system. In recent centuries it more often means a prescribed process for completing a task. It may refer to: *Scientific method, a series of steps, or collection of methods, taken to acquire knowledge *Method (computer programming), a piece of code associated with a class or object to perform a task * Method (patent), under patent law, a protected series of steps or acts *Methodology, comparison or study and critique of individual methods that are used in a given discipline or field of inquiry *''Discourse on the Method'', a philosophical and mathematical treatise by René Descartes * ''Methods'' (journal), a scientific journal covering research on techniques in the experimental biological and medical sciences Arts *Method (music), a kind of textbook to help students learning to play a musical instrument * ''Method'' (2004 film), a 2004 film directed b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplotype
A haplotype ( haploid genotype) is a group of alleles in an organism that are inherited together from a single parent. Many organisms contain genetic material ( DNA) which is inherited from two parents. Normally these organisms have their DNA organized in two sets of pairwise similar chromosomes. The offspring gets one chromosome in each pair from each parent. A set of pairs of chromosomes is called diploid and a set of only one half of each pair is called haploid. The haploid genotype (haplotype) is a genotype that considers the singular chromosomes rather than the pairs of chromosomes. It can be all the chromosomes from one of the parents or a minor part of a chromosome, for example a sequence of 9000 base pairs. However, there are other uses of this term. First, it is used to mean a collection of specific alleles (that is, specific DNA sequences) in a cluster of tightly linked genes on a chromosome that are likely to be inherited together—that is, they are likely to be co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
International HapMap Project
The International HapMap Project was an organization that aimed to develop a haplotype map (HapMap) of the human genome, to describe the common patterns of human genetic variation. HapMap is used to find genetic variants affecting health, disease and responses to drugs and environmental factors. The information produced by the project is made freely available for research. The International HapMap Project is a collaboration among researchers at academic centers, non-profit biomedical research groups and private companies in Canada, China (including Hong Kong), Japan, Nigeria, the United Kingdom, and the United States. It officially started with a meeting on October 27 to 29, 2002, and was expected to take about three years. It comprises two phases; the complete data obtained in Phase I were published on 27 October 2005. The analysis of the Phase II dataset was published in October 2007. The Phase III dataset was released in spring 2009 and the publication presenting the final re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biobanks
A biobank is a type of biorepository that stores biological samples (usually human) for use in research. Biobanks have become an important resource in medical research, supporting many types of contemporary research like genomics and personalized medicine. Biobanks can give researchers access to data representing a large number of people. Samples in biobanks and the data derived from those samples can often be used by multiple researchers for cross purpose research studies. For example, many diseases are associated with single-nucleotide polymorphisms. Genome-wide association studies using data from tens or hundreds of thousands of individuals can identify these genetic associations as potential disease biomarkers. Many researchers struggled to acquire sufficient samples prior to the advent of biobanks. Biobanks have provoked questions on privacy, research ethics, and medical ethics. Viewpoints on what constitutes appropriate biobank ethics diverge. However, a consensus has been ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Association
Genetic association is when one or more genotypes within a population co-occur with a phenotypic trait more often than would be expected by chance occurrence. Studies of genetic association aim to test whether single-locus alleles or genotype frequencies (or more generally, multilocus haplotype frequencies) differ between two groups of individuals (usually diseased subjects and healthy controls). Genetic association studies today are based on the principle that genotypes can be compared "directly", i.e. with the sequences of the actual genomes or exomes via whole genome sequencing or whole exome sequencing. Before 2010, DNA sequencing methods were used. Description Genetic association can be between phenotypes, such as visible characteristics such as flower color or height, between a phenotype and a genetic polymorphism, such as a single nucleotide polymorphism (SNP), or between two genetic polymorphisms. Association between genetic polymorphisms occurs when there is non-r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |