|
FoldX
FoldX is a protein design algorithm that uses an empirical force field. It can determine the energetic effect of point mutations as well as the interaction energy of protein complexes (including Protein- DNA). FoldX can mutate protein and DNA side chains using a probability-based rotamer library, while exploring alternative conformations of the surrounding side chains. Applications * Prediction of the effect of point mutations or human SNPs on protein stability or protein complexes * Protein design to improve stability or modify affinity or specificity * Homology modeling The FoldX force field The energy function includes terms that have been found to be important for protein stability, where the energy of unfolding (∆G) of a target protein is calculated using the equation: ∆G = ∆Gvdw + ∆GsolvH + ∆GsolvP + ∆Ghbond + ∆Gwb + ∆Gel + ∆Smc + ∆Ssc Where ∆Gvdw is the sum of the Van der Waals contributions of all atoms with respect to the same interaction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Design Software
Molecular design software is notable software for molecular modeling, that provides special support for developing molecular models ''de novo''. In contrast to the usual molecular modeling programs, such as for molecular dynamics and quantum chemistry, such software ''directly'' supports the aspects related to constructing molecular models, including: * Molecular graphics * interactive molecular drawing and conformational editing * building polymeric molecules, crystals, and solvated systems * partial charges development * geometry optimization * support for the different aspects of force field development Comparison of software covering the major aspects of molecular design Notes and references See also {{columns-list, colwidth=30em, *Molecule editor *Molecular modelling *Molecular modeling on GPUs *Protein design *Drug design *Force field (chemistry) *Comparison of force field implementations *Comparison of nucleic acid simulation software *Comparison of software for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Command Line
A command-line interpreter or command-line processor uses a command-line interface (CLI) to receive commands from a user in the form of lines of text. This provides a means of setting parameters for the environment, invoking executables and providing information to them as to what actions they are to perform. In some cases the invocation is conditional based on conditions established by the user or previous executables. Such access was first provided by computer terminals starting in the mid-1960s. This provided an interactive environment not available with punched cards or other input methods. Today, many users rely upon graphical user interfaces and menu-driven interactions. However, some programming and maintenance tasks may not have a graphical user interface and use a command line. Alternatives to the command-line interface include text-based user interface menus (for example, IBM AIX SMIT), keyboard shortcuts, and various desktop metaphors centered on the pointer (usual ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
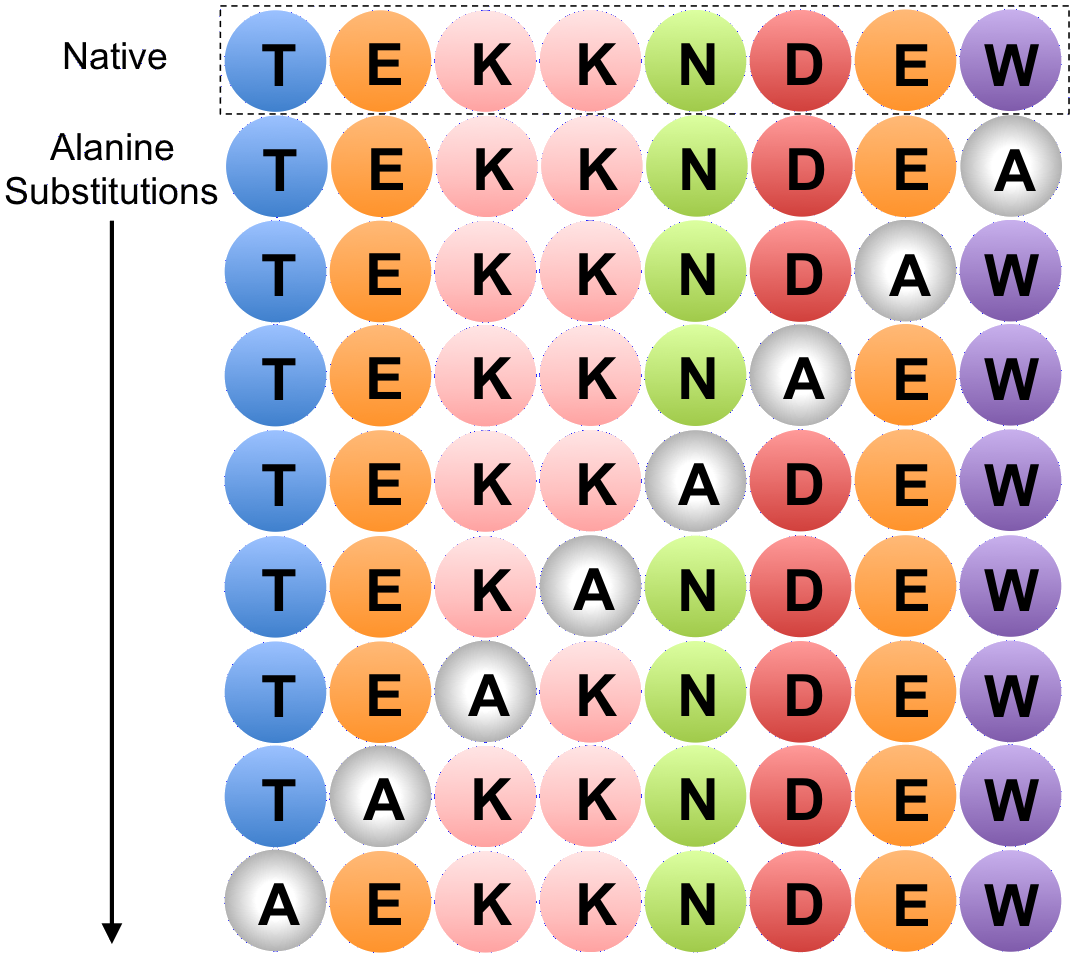
Alanine Scan
In molecular biology, alanine scanning is a site-directed mutagenesis technique used to determine the contribution of a specific residue to the stability or function of a given protein. Alanine is used because of its non-bulky, chemically inert, methyl functional group that nevertheless mimics the secondary structure preferences that many of the other amino acids possess. Sometimes bulky amino acids such as valine or leucine are used in cases where conservation of the size of mutated residues is needed. This technique can also be used to determine whether the side chain of a specific residue plays a significant role in bioactivity. This is usually accomplished by site-directed mutagenesis or randomly by creating a PCR library. Furthermore, computational methods to estimate thermodynamic parameters based on theoretical alanine substitutions have been developed. This technique is rapid, because many side chains are analyzed simultaneously and the need for protein purification and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Site-directed Mutagenesis
Site-directed mutagenesis is a molecular biology method that is used to make specific and intentional mutating changes to the DNA sequence of a gene and any gene products. Also called site-specific mutagenesis or oligonucleotide-directed mutagenesis, it is used for investigating the structure and biological activity of DNA, RNA, and protein molecules, and for protein engineering. Site-directed mutagenesis is one of the most important laboratory techniques for creating DNA libraries by introducing mutations into DNA sequences. There are numerous methods for achieving site-directed mutagenesis, but with decreasing costs of oligonucleotide synthesis, artificial gene synthesis is now occasionally used as an alternative to site-directed mutagenesis. Since 2013, the development of the CRISPR/Cas9 technology, based on a prokaryotic viral defense system, has also allowed for the editing of the genome, and mutagenesis may be performed ''in vivo'' with relative ease. History Early attempt ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a ''residue'' indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Energy Minimization
In the field of computational chemistry, energy minimization (also called energy optimization, geometry minimization, or geometry optimization) is the process of finding an arrangement in space of a collection of atoms where, according to some computational model of chemical bonding, the net inter-atomic force on each atom is acceptably close to zero and the position on the potential energy surface (PES) is a stationary point (described later). The collection of atoms might be a single molecule, an ion, a condensed phase, a transition state or even a collection of any of these. The computational model of chemical bonding might, for example, be quantum mechanics. As an example, when optimizing the geometry of a water molecule, one aims to obtain the hydrogen-oxygen bond lengths and the hydrogen-oxygen-hydrogen bond angle which minimize the forces that would otherwise be pulling atoms together or pushing them apart. The motivation for performing a geometry optimization is the physic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Entropy
Entropy is a scientific concept, as well as a measurable physical property, that is most commonly associated with a state of disorder, randomness, or uncertainty. The term and the concept are used in diverse fields, from classical thermodynamics, where it was first recognized, to the microscopic description of nature in statistical physics, and to the principles of information theory. It has found far-ranging applications in chemistry and physics, in biological systems and their relation to life, in cosmology, economics, sociology, weather science, climate change, and information systems including the transmission of information in telecommunication. The thermodynamic concept was referred to by Scottish scientist and engineer William Rankine in 1850 with the names ''thermodynamic function'' and ''heat-potential''. In 1865, German physicist Rudolf Clausius, one of the leading founders of the field of thermodynamics, defined it as the quotient of an infinitesimal amount of hea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogen Bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a lone pair of electrons—the hydrogen bond acceptor (Ac). Such an interacting system is generally denoted , where the solid line denotes a polar covalent bond, and the dotted or dashed line indicates the hydrogen bond. The most frequent donor and acceptor atoms are the second-row elements nitrogen (N), oxygen (O), and fluorine (F). Hydrogen bonds can be intermolecular (occurring between separate molecules) or intramolecular (occurring among parts of the same molecule). The energy of a hydrogen bond depends on the geometry, the environment, and the nature of the specific donor and acceptor atoms and can vary between 1 and 40 kcal/mol. This makes them somewhat stronger than a van der Waals interaction, and weaker than fully covalent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Force Field (chemistry)
In the context of chemistry and molecular modelling, a force field is a computational method that is used to estimate the forces between atoms within molecules and also between molecules. More precisely, the force field refers to the functional form and parameter sets used to calculate the potential energy of a system of atoms or coarse-grained particles in molecular mechanics, molecular dynamics, or Monte Carlo simulations. The parameters for a chosen energy function may be derived from experiments in physics and chemistry, calculations in quantum mechanics, or both. Force fields are interatomic potentials and utilize the same concept as force fields in classical physics, with the difference that the force field parameters in chemistry describe the energy landscape, from which the acting forces on every particle are derived as a gradient of the potential energy with respect to the particle coordinates. ''All-atom'' force fields provide parameters for every type of atom in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |